Implementing tumor mutational burden (TMB) analysis in routine diagnostics-a primer for molecular pathologists and clinicians - PubMed (original) (raw)
Review
doi: 10.21037/tlcr.2018.08.14.
Jan Budczies 1 2, Petros Christopoulos 3 4, Volker Endris 1, Amelie Lier 1, Eugen Rempel 1, Anna-Lena Volckmar 1, Martina Kirchner 1, Moritz von Winterfeld 1, Jonas Leichsenring 1, Olaf Neumann 1, Stefan Fröhling 5 6, Roland Penzel 1, Michael Thomas 3 4, Peter Schirmacher 1 2, Albrecht Stenzinger 1 2
Affiliations
- PMID: 30505715
- PMCID: PMC6249620
- DOI: 10.21037/tlcr.2018.08.14
Review
Implementing tumor mutational burden (TMB) analysis in routine diagnostics-a primer for molecular pathologists and clinicians
Michael Allgäuer et al. Transl Lung Cancer Res. 2018 Dec.
Abstract
Tumor mutational burden (TMB) is a new biomarker for prediction of response to PD-(L)1 treatment. Comprehensive sequencing approaches (i.e., whole exome and whole genome sequencing) are ideally suited to measure TMB directly. However, as their applicability in routine diagnostics is currently limited by high costs, long turnaround times and poor availability of fresh tissue, targeted next-generation sequencing (NGS) of formalin-fixed and paraffin-embedded (FFPE) samples appears to be a more feasible and straight-forward approach for TMB approximation, which can be seamlessly integrated in already existing diagnostic workflows and pipelines. In this work, we provide an overview of the clinical implications of TMB testing and highlight key parameters including pre-analysis, analysis and post-analytical steps that influence and shape TMB approximation by panel sequencing. Collectively, the data will not only serve as a field guide and state of the art knowledge source for molecular pathologists who consider implementation of TMB measurement in their lab, but also enable clinicians in understanding the specific parameters influencing TMB test results and reporting.
Keywords: Tumor mutational burden (TMB); mutational load; next-generation sequencing (NGS); panel; sequencing.
Conflict of interest statement
Conflicts of Interest: V Endris: advisory board and lecture fees from AstraZeneca and ThermoFisher. J Leichsenring: consultancy contract with AstraZeneca. S Fröhling: speaker’s honoraria from Amgen, Lilly, PharmaMar and Roche; research funding from AstraZeneca, Pfizer and PharmaMar. M Thomas: advisory board honoraria from Novartis, Lilly, BMS, MSD, Roche, Celgene, Takeda, AbbVie, Boehringer, speaker’s honoraria from Lilly, MSD, Takeda, research funding from AstraZeneca, BMS, Celgene, Novartis, Roche and travel grants from BMS, MSD, Novartis, Boehringer. P Schirmacher: advisory board honoraria from Pfizer, Roche, Novartis, AstraZeneca as well as speaker’s honoraria and research funding from Roche, AstraZeneca and Novartis. A Stenzinger: advisory board honoraria from BMS, AstraZeneca, Novartis, ThermoFisher, speaker’s honoraria from BMS, Illumina, AstraZeneca, MSD, Novartis, Roche, ThermoFisher, and research funding from Chugai. The other authors have no conflicts of interest to declare.
Figures
Figure 1
One-stop shop approach to maximize specimen yield. Necessary molecular testing should best be indicated by the clinician or anticipated when the specimen is initially processed in the pathology laboratory. A sufficient number of slides should be precut to avoid re-cutting of the tissue block. Depicted are three groups of diagnostic tests that are often performed sequentially. # slides: approximated number of slides needed. In routine pathology, material usage is determined by the utilization of IHC stains. In molecular pathology, the number of slides/paraffin sections needed depends on the amount of tumor present and assays used.
Figure 2
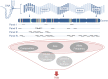
Panel design influences TMB measurement. Amplicons (i.e., sequenced regions of exome) of arbitrary sequencing panels (Panels I–IV) are schematically depicted to illustrate differences in size and composition. Panel I is a small focused panel which might be used for entity specific investigations or when DNA is limited, like in liquid biopsies. Panel II and III are more comprehensive targeting additional exonic regions. Panel IV is a comprehensive tumor profiling panel developed for TMB detection. Indicated on exome are exemplified clonal (green), subclonal (red), and frameshift (orange) mutations, and indels (blue). TMB, tumor mutational burden.
Figure 3
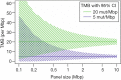
Precision of TMB estimation using targeted sequencing panels of size 0.1 to 10 Mbp. An upper limit for the precision of TMB estimates is set by the combinatorial error that comes from estimating the mutation rate (in mut/Mbp) by the number of mutations in a sequence of a limited length. In the display, the precision of TMB estimation (reported as 95% confidence interval) is illustrated for a tumor with high TMB (20 mut/Mbp, typically classified as immune therapy responder) and a tumor with low TMB (5 mut/Mbp, typically classified as immune therapy non-responder). Sequencing panels larger than 1 Mbp are required to separate the tumors in the example with high precision. TMB, tumor mutational burden.
Similar articles
- Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels.
Endris V, Buchhalter I, Allgäuer M, Rempel E, Lier A, Volckmar AL, Kirchner M, von Winterfeld M, Leichsenring J, Neumann O, Penzel R, Weichert W, Glimm H, Fröhling S, Winter H, Herth F, Thomas M, Schirmacher P, Budczies J, Stenzinger A. Endris V, et al. Int J Cancer. 2019 May 1;144(9):2303-2312. doi: 10.1002/ijc.32002. Epub 2019 Feb 4. Int J Cancer. 2019. PMID: 30446996 - Size matters: Dissecting key parameters for panel-based tumor mutational burden analysis.
Buchhalter I, Rempel E, Endris V, Allgäuer M, Neumann O, Volckmar AL, Kirchner M, Leichsenring J, Lier A, von Winterfeld M, Penzel R, Christopoulos P, Thomas M, Fröhling S, Schirmacher P, Budczies J, Stenzinger A. Buchhalter I, et al. Int J Cancer. 2019 Feb 15;144(4):848-858. doi: 10.1002/ijc.31878. Epub 2018 Dec 4. Int J Cancer. 2019. PMID: 30238975 - Association Between Preanalytical Factors and Tumor Mutational Burden Estimated by Next-Generation Sequencing-Based Multiplex Gene Panel Assay.
Quy PN, Kanai M, Fukuyama K, Kou T, Kondo T, Yamamoto Y, Matsubara J, Hiroshima A, Mochizuki H, Sakuma T, Kamada M, Nakatsui M, Eso Y, Seno H, Masui T, Takaori K, Minamiguchi S, Matsumoto S, Muto M. Quy PN, et al. Oncologist. 2019 Dec;24(12):e1401-e1408. doi: 10.1634/theoncologist.2018-0587. Epub 2019 Jun 11. Oncologist. 2019. PMID: 31186376 Free PMC article. - Tumor mutational burden standardization initiatives: Recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions.
Stenzinger A, Allen JD, Maas J, Stewart MD, Merino DM, Wempe MM, Dietel M. Stenzinger A, et al. Genes Chromosomes Cancer. 2019 Aug;58(8):578-588. doi: 10.1002/gcc.22733. Epub 2019 Mar 7. Genes Chromosomes Cancer. 2019. PMID: 30664300 Free PMC article. Review. - Methods of measurement for tumor mutational burden in tumor tissue.
Meléndez B, Van Campenhout C, Rorive S, Remmelink M, Salmon I, D'Haene N. Meléndez B, et al. Transl Lung Cancer Res. 2018 Dec;7(6):661-667. doi: 10.21037/tlcr.2018.08.02. Transl Lung Cancer Res. 2018. PMID: 30505710 Free PMC article. Review.
Cited by
- Prognostic model for hepatocellular carcinoma based on necroptosis-related genes and analysis of drug treatment responses.
Wu R, Deng X, Wang X, Li S, Su J, Sun X. Wu R, et al. Heliyon. 2024 Aug 22;10(17):e36561. doi: 10.1016/j.heliyon.2024.e36561. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39263127 Free PMC article. - Low expression of ACOT13 predicts poor prognosis and immunotherapy outcome in ovarian cancer.
Xie T, Liu T, Wu L, Yi N. Xie T, et al. Am J Transl Res. 2024 Apr 15;16(4):1424-1441. doi: 10.62347/OIQB4997. eCollection 2024. Am J Transl Res. 2024. PMID: 38715807 Free PMC article. - Identification of AURKA as a Biomarker Associated with Cuproptosis and Ferroptosis in HNSCC.
Jia X, Tian J, Fu Y, Wang Y, Yang Y, Zhang M, Yang C, Liu Y. Jia X, et al. Int J Mol Sci. 2024 Apr 16;25(8):4372. doi: 10.3390/ijms25084372. Int J Mol Sci. 2024. PMID: 38673957 Free PMC article. - TMBcalc: a computational pipeline for identifying pan-cancer Tumor Mutational Burden gene signatures.
Privitera GF, Alaimo S, Caruso A, Ferro A, Forte S, Pulvirenti A. Privitera GF, et al. Front Genet. 2024 Apr 5;15:1285305. doi: 10.3389/fgene.2024.1285305. eCollection 2024. Front Genet. 2024. PMID: 38645485 Free PMC article. - Electrochemical determination of rutin by using NiFe2O4 nanoparticles-loaded reduced graphene oxide.
Askari N, Salarizadeh N, Askari MB. Askari N, et al. J Mater Sci Mater Electron. 2021;32(8):9765-9775. doi: 10.1007/s10854-021-05636-9. Epub 2021 Mar 12. J Mater Sci Mater Electron. 2021. PMID: 38624849 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous