The Genome of the North American Brown Bear or Grizzly: Ursus arctos ssp. horribilis - PubMed (original) (raw)
doi: 10.3390/genes9120598.
Heather Kirk 2, Lauren Coombe 3, Shaun D Jackman 4, Justin Chu 5, Kane Tse 6, Dean Cheng 7, Eric Chuah 8, Pawan Pandoh 9, Rebecca Carlsen 10, Yongjun Zhao 11, Andrew J Mungall 12, Richard Moore 13, Inanc Birol 14 15, Maria Franke 16, Marco A Marra 17 18, Christopher Dutton 19, Steven J M Jones 20 21 22
Affiliations
- PMID: 30513700
- PMCID: PMC6315469
- DOI: 10.3390/genes9120598
The Genome of the North American Brown Bear or Grizzly: Ursus arctos ssp. horribilis
Gregory A Taylor et al. Genes (Basel). 2018.
Abstract
The grizzly bear (Ursus arctos ssp. horribilis) represents the largest population of brown bears in North America. Its genome was sequenced using a microfluidic partitioning library construction technique, and these data were supplemented with sequencing from a nanopore-based long read platform. The final assembly was 2.33 Gb with a scaffold N50 of 36.7 Mb, and the genome is of comparable size to that of its close relative the polar bear (2.30 Gb). An analysis using 4104 highly conserved mammalian genes indicated that 96.1% were found to be complete within the assembly. An automated annotation of the genome identified 19,848 protein coding genes. Our study shows that the combination of the two sequencing modalities that we used is sufficient for the construction of highly contiguous reference quality mammalian genomes. The assembled genome sequence and the supporting raw sequence reads are available from the NCBI (National Center for Biotechnology Information) under the bioproject identifier PRJNA493656, and the assembly described in this paper is version QXTK01000000.
Keywords: Ursus arctos ssp. Horribilis; genome; grizzly bear; microfluidic partitioning; nanopore.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
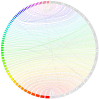
Figure 1
Jupiter plot. A global genome alignment, using BWA-MEM, of the grizzly genome (left side of circle) to the polar bear genome (right side). Connections show the aligned regions of each assembly. The grizzly scaffolds are limited to those over 10 Mb in length (>85% of the assembly). The longest polar bear scaffolds were selected to sum to the same amount of sequence (2 Gb). Only alignments over 10 kb in length are displayed.
Similar articles
- First report of Taenia arctos (Cestoda: Taeniidae) from grizzly (Ursus arctos horribilis) and black bears (Ursus americanus) in North America.
Catalano S, Lejeune M, Verocai GG, Duignan PJ. Catalano S, et al. Parasitol Int. 2014 Apr;63(2):389-91. doi: 10.1016/j.parint.2013.12.012. Epub 2013 Dec 29. Parasitol Int. 2014. PMID: 24382413 - INVESTIGATION OF SARCOCYSTIS SPP. INFECTION IN FREE-RANGING AMERICAN BLACK BEARS (URSUS AMERICANUS) AND GRIZZLY BEARS (URSUS ARCTOS HORRIBILIS) IN BRITISH COLUMBIA, CANADA.
Lee LKF, McGregor GF, Haman KH, Raverty S, Grigg ME, Shapiro K, Schwantje H, Schofer D, Lee MJ, Himsworth CG, Byers KA. Lee LKF, et al. J Wildl Dis. 2021 Oct 1;57(4):856-864. doi: 10.7589/JWD-D-20-00225. J Wildl Dis. 2021. PMID: 34516653 - A Beary Good Genome: Haplotype-Resolved, Chromosome-Level Assembly of the Brown Bear (Ursus arctos).
Armstrong EE, Perry BW, Huang Y, Garimella KV, Jansen HT, Robbins CT, Tucker NR, Kelley JL. Armstrong EE, et al. Genome Biol Evol. 2022 Sep 6;14(9):evac125. doi: 10.1093/gbe/evac125. Genome Biol Evol. 2022. PMID: 35929770 Free PMC article. - Conservation and management of the culture of bears.
Servheen C, Gunther KA. Servheen C, et al. Ecol Evol. 2022 Apr 19;12(4):e8840. doi: 10.1002/ece3.8840. eCollection 2022 Apr. Ecol Evol. 2022. PMID: 35462976 Free PMC article. Review. - Functional macronutritional generalism in a large omnivore, the brown bear.
Coogan SCP, Raubenheimer D, Stenhouse GB, Coops NC, Nielsen SE. Coogan SCP, et al. Ecol Evol. 2018 Jan 29;8(4):2365-2376. doi: 10.1002/ece3.3867. eCollection 2018 Feb. Ecol Evol. 2018. PMID: 29468050 Free PMC article. Review.
Cited by
- Examining the Effects of Hibernation on Germline Mutation Rates in Grizzly Bears.
Wang RJ, Peña-Garcia Y, Bibby MG, Raveendran M, Harris RA, Jansen HT, Robbins CT, Rogers J, Kelley JL, Hahn MW. Wang RJ, et al. Genome Biol Evol. 2022 Oct 7;14(10):evac148. doi: 10.1093/gbe/evac148. Genome Biol Evol. 2022. PMID: 36173788 Free PMC article. - A genome assembly of the Atlantic chub mackerel (Scomber colias): a valuable teleost fishing resource.
Machado AM, Gomes-Dos-Santos A, Fonseca MM, da Fonseca RR, Veríssimo A, Felício M, Capela R, Alves N, Santos M, Salvador-Caramelo F, Domingues M, Ruivo R, Froufe E, Castro LFC. Machado AM, et al. GigaByte. 2022 Feb 14;2022:gigabyte40. doi: 10.46471/gigabyte.40. eCollection 2022. GigaByte. 2022. PMID: 36824513 Free PMC article. - Analyses of key genes involved in Arctic adaptation in polar bears suggest selection on both standing variation and de novo mutations played an important role.
Samaniego Castruita JA, Westbury MV, Lorenzen ED. Samaniego Castruita JA, et al. BMC Genomics. 2020 Aug 6;21(1):543. doi: 10.1186/s12864-020-06940-0. BMC Genomics. 2020. PMID: 32758141 Free PMC article. - The genome of the forest insect pest Pissodes strobi reveals genome expansion and evidence of a Wolbachia endosymbiont.
Gagalova KK, Whitehill JGA, Culibrk L, Lin D, Lévesque-Tremblay V, Keeling CI, Coombe L, Yuen MMS, Birol I, Bohlmann J, Jones SJM. Gagalova KK, et al. G3 (Bethesda). 2022 Apr 4;12(4):jkac038. doi: 10.1093/g3journal/jkac038. G3 (Bethesda). 2022. PMID: 35171977 Free PMC article. - Chromosome-Level Reference Genome Assembly for the American Pika (Ochotona princeps).
Sjodin BMF, Galbreath KE, Lanier HC, Russello MA. Sjodin BMF, et al. J Hered. 2021 Nov 1;112(6):549-557. doi: 10.1093/jhered/esab031. J Hered. 2021. PMID: 34036348 Free PMC article.
References
- Gyug L., Hamilton T., Austin M. Grizzly bear (Ursus Arctos). Accounts and Measures for Managing Identified Wildlife–Accounts V2004. Ministry of Water, Land and Air Protection British Columbia: National Library of Canada; BC, Canada: 2004.
LinkOut - more resources
Full Text Sources
Miscellaneous