Global Proteomic Profiling of Salmonella Infection by a Giant Phage - PubMed (original) (raw)
. 2019 Feb 19;93(5):e01833-18.
doi: 10.1128/JVI.01833-18. Print 2019 Mar 1.
Nurul Humaira Mohd Redzuan 2, Melissa K Barton 2, Nur Amira Md Amin 2, Maxim I Desmond 2, Lily E Adams 2, Bazla Ali 2, Sammy Pardo 1, Dana Molleur 1, Weimin Wu 3, William W Newcomb 3, Michael V Osier 2, Lindsay W Black 4, Alasdair C Steven 3, Julie A Thomas 5
Affiliations
- PMID: 30541839
- PMCID: PMC6384053
- DOI: 10.1128/JVI.01833-18
Global Proteomic Profiling of Salmonella Infection by a Giant Phage
Susan T Weintraub et al. J Virol. 2019.
Abstract
The 240-kb Salmonella phage SPN3US genome encodes 264 gene products, many of which are functionally uncharacterized. We have previously used mass spectrometry to define the proteomes of wild-type and mutant forms of the SPN3US virion. In this study, we sought to determine whether this technique was suitable for the characterization of the SPN3US proteome during liquid infection. Mass spectrometry of SPN3US-infected cells identified 232 SPN3US and 1,994 Salmonella proteins. SPN3US proteins with related functions, such as proteins with roles in DNA replication, transcription, and virion formation, were coordinately expressed in a temporal manner. Mass spectral counts showed the four most abundant SPN3US proteins to be the major capsid protein, two head ejection proteins, and the functionally unassigned protein gp22. This high abundance of gp22 in infected bacteria contrasted with its absence from mature virions, suggesting that it might be the scaffold protein, an essential head morphogenesis protein yet to be identified in giant phages. We identified homologs to SPN3US gp22 in 45 related giant phages, including ϕKZ, whose counterpart is also abundant in infected bacteria but absent in the virion. We determined the ϕKZ counterpart to be cleaved in vitro by its prohead protease, an event that has been observed to promote head maturation of some other phages. Our findings are consistent with a scaffold protein assignment for SPN3US gp22, although direct evidence is required for its confirmation. These studies demonstrate the power of mass spectral analyses for facilitating the acquisition of new knowledge into the molecular events of viral infection.IMPORTANCE "Giant" phages with genomes >200 kb are being isolated in increasing numbers from a range of environments. With hosts such as Salmonella enterica, Pseudomonas aeruginosa, and Erwinia amylovora, these phages are of interest for phage therapy of multidrug-resistant pathogens. However, our understanding of how these complex phages interact with their hosts is impeded by the proportion (∼80%) of their gene products that are functionally uncharacterized. To develop the repertoire of techniques for analysis of phages, we analyzed a liquid infection of Salmonella phage SPN3US (240-kb genome) using third-generation mass spectrometry. We observed the temporal production of phage proteins whose genes collectively represent 96% of the SPN3US genome. These findings demonstrate the sensitivity of mass spectrometry for global proteomic profiling of virus-infected cells, and the identification of a candidate for a major head morphogenesis protein will facilitate further studies into giant phage head assembly.
Keywords: Salmonella; bacteriophage assembly; giant phage; mass spectrometry.
Figures
FIG 1
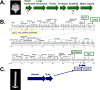
Major genes required to form the virion of a myovirus (phage with a contractile tail). (A) Scheme of the ordering of genes required to make the head (or capsid) of most tailed phages. Red question marks indicate genes that have yet to be identified in SPN3US. (B) Linear representation of the SPN3US genome showing the locations of known head and tail morphogenesis genes from panel A. (C) Representation of the major genes required to make a contractile tail. Capsid and tail images are by W. Wu (work in progress).
FIG 2
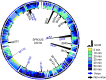
Production of SPN3US proteins during liquid infection of Salmonella enterica serovar Typhimurium LT2. Black inner ring peaks represent an estimate of relative abundance (SC/M, spectral count divided by molecular mass) of each protein at Time-60. The relative numbers of mass spectral counts at each time point adjusted by the highest number of spectra detected for each protein is indicated. The exterior ring depicts the SPN3US genome map with virion protein genes shaded blue and nonvirion protein genes shaded black. Highly abundant proteins, including major virion proteins, the major capsid protein (MCP), the tail sheath, and tube proteins are labeled.
FIG 3
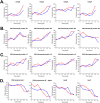
Identification of Salmonella proteins during infection by SPN3US, and in an uninfected control. (A) Major outer membrane proteins. (B and C) Major ribosomal proteins and proteins with roles relating to protein expression and folding. (D) Major RNA polymerase and DNA polymerase subunits.
FIG 4
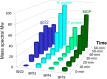
Mass spectral identification of major SPN3US major head proteins and the scaffold protein candidate, gp22, during infection.
FIG 5
The gene locale of SPN3US scaffold candidate, gp22, is conserved in related phages.
FIG 6
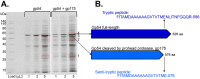
Assay of ϕKZ gp54 with the prohead protease gp175. (A) SDS-PAGE gel of assay of recombinant ϕKZ proteins gp54 with gp175 in a lysate of E. coli proteins. A control of gp54 only was included. The amounts of each sample loaded are indicated. Gel slices processed for MS analyses are indicated by a red square. (B) Semitryptic and tryptic mass spectra for gp54 identified by MS in the gel slices indicated in panel A.
Similar articles
- A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid.
Reilly ER, Abajorga MK, Kiser C, Mohd Redzuan NH, Haidar Z, Adams LE, Diaz R, Pinzon JA, Hudson AO, Black LW, Hsia RC, Weintraub ST, Thomas JA. Reilly ER, et al. Viruses. 2020 Jul 5;12(7):725. doi: 10.3390/v12070725. Viruses. 2020. PMID: 32635654 Free PMC article. - The Mottled Capsid of the Salmonella Giant Phage SPN3US, a Likely Maturation Intermediate with a Novel Internal Shell.
Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC. Heymann JB, et al. Viruses. 2020 Aug 19;12(9):910. doi: 10.3390/v12090910. Viruses. 2020. PMID: 32825132 Free PMC article. - Identification of Essential Genes in the Salmonella Phage SPN3US Reveals Novel Insights into Giant Phage Head Structure and Assembly.
Thomas JA, Benítez Quintana AD, Bosch MA, Coll De Peña A, Aguilera E, Coulibaly A, Wu W, Osier MV, Hudson AO, Weintraub ST, Black LW. Thomas JA, et al. J Virol. 2016 Oct 28;90(22):10284-10298. doi: 10.1128/JVI.01492-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27605673 Free PMC article. - To Be or Not To Be T4: Evidence of a Complex Evolutionary Pathway of Head Structure and Assembly in Giant Salmonella Virus SPN3US.
Ali B, Desmond MI, Mallory SA, Benítez AD, Buckley LJ, Weintraub ST, Osier MV, Black LW, Thomas JA. Ali B, et al. Front Microbiol. 2017 Nov 15;8:2251. doi: 10.3389/fmicb.2017.02251. eCollection 2017. Front Microbiol. 2017. PMID: 29187846 Free PMC article. - Structure and physiology of giant DNA viruses.
Dos Santos Oliveira J, Lavell AA, Essus VA, Souza G, Nunes GHP, Benício E, Guimarães AJ, Parent KN, Cortines JR. Dos Santos Oliveira J, et al. Curr Opin Virol. 2021 Aug;49:58-67. doi: 10.1016/j.coviro.2021.04.012. Epub 2021 May 26. Curr Opin Virol. 2021. PMID: 34051592 Review.
Cited by
- A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid.
Reilly ER, Abajorga MK, Kiser C, Mohd Redzuan NH, Haidar Z, Adams LE, Diaz R, Pinzon JA, Hudson AO, Black LW, Hsia RC, Weintraub ST, Thomas JA. Reilly ER, et al. Viruses. 2020 Jul 5;12(7):725. doi: 10.3390/v12070725. Viruses. 2020. PMID: 32635654 Free PMC article. - The Mottled Capsid of the Salmonella Giant Phage SPN3US, a Likely Maturation Intermediate with a Novel Internal Shell.
Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC. Heymann JB, et al. Viruses. 2020 Aug 19;12(9):910. doi: 10.3390/v12090910. Viruses. 2020. PMID: 32825132 Free PMC article. - Molecular Characterization and Comparative Genomic Analysis of vB_PaeP_YA3, a Novel Temperate Bacteriophage of Pseudomonas aeruginosa.
Yu X, Xu J, Gu Y, Zhang R, Zhu Y, Liu X. Yu X, et al. Front Microbiol. 2020 Jun 3;11:947. doi: 10.3389/fmicb.2020.00947. eCollection 2020. Front Microbiol. 2020. PMID: 32655502 Free PMC article. - Genomic Characterization of Jumbo Salmonella Phages That Effectively Target United Kingdom Pig-Associated Salmonella Serotypes.
Thanki AM, Brown N, Millard AD, Clokie MRJ. Thanki AM, et al. Front Microbiol. 2019 Jul 2;10:1491. doi: 10.3389/fmicb.2019.01491. eCollection 2019. Front Microbiol. 2019. PMID: 31312191 Free PMC article. - Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Armbruster E, McCammon JA, Amaro RE, Pogliano J, Corbett KD, Villa E. Laughlin TG, et al. Nature. 2022 Aug;608(7922):429-435. doi: 10.1038/s41586-022-05013-4. Epub 2022 Aug 3. Nature. 2022. PMID: 35922510 Free PMC article.
References
- Hendrix RW. 2009. Jumbo bacteriophages, p 229–240. In Van Etten J. (ed), Lesser known large dsDNA viruses, vol 328 Springer, Berlin, Germany. - PubMed
- Serwer P, Hayes SJ, Thomas J, Demeler B, Hardies SC. 2009. Isolation of novel large and aggregating bacteriophages, p 55–66. In Clokie M, Kropinski AM (ed), Bacteriophages: methods and protocols. Humana Press, New York, NY. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI011676/AI/NIAID NIH HHS/United States
- R01 GM118766/GM/NIGMS NIH HHS/United States
- S10 RR025111/RR/NCRR NIH HHS/United States
- UA5 GM126533/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases