Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora - PubMed (original) (raw)
Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora
Huei-Jiun Su et al. Proc Natl Acad Sci U S A. 2019.
Abstract
Plastid genomes (plastomes) vary enormously in size and gene content among the many lineages of nonphotosynthetic plants, but key lineages remain unexplored. We therefore investigated plastome sequence and expression in the holoparasitic and morphologically bizarre Balanophoraceae. The two Balanophora plastomes examined are remarkable, exhibiting features rarely if ever seen before in plastomes or in any other genomes. At 15.5 kb in size and with only 19 genes, they are among the most reduced plastomes known. They have no tRNA genes for protein synthesis, a trait found in only three other plastid lineages, and thus Balanophora plastids must import all tRNAs needed for translation. Balanophora plastomes are exceptionally compact, with numerous overlapping genes, highly reduced spacers, loss of all _cis_-spliced introns, and shrunken protein genes. With A+T contents of 87.8% and 88.4%, the Balanophora genomes are the most AT-rich genomes known save for a single mitochondrial genome that is merely bloated with AT-rich spacer DNA. Most plastid protein genes in Balanophora consist of ≥90% AT, with several between 95% and 98% AT, resulting in the most biased codon usage in any genome described to date. A potential consequence of its radical compositional evolution is the novel genetic code used by Balanophora plastids, in which TAG has been reassigned from stop to tryptophan. Despite its many exceptional properties, the Balanophora plastome must be functional because all examined genes are transcribed, its only intron is correctly _trans_-spliced, and its protein genes, although highly divergent, are evolving under various degrees of selective constraint.
Keywords: AT-biased base composition; genetic code change; genome reduction; overlapping genes; parasitic plants.
Conflict of interest statement
Conflict of interest statement: E.K.W. and C.W.d. are coauthors on a 2018 paper with reviewer I.D.S. This paper had 42 authors with E.K.W., C.W.d., and I.D.S. all middle authors. E.K.W. and C.W.d. had no significant interaction with I.D.S. on the project or paper.
Figures
Fig. 1.
Morphology and microscopy of Balanophora. Male (A) and female (B) plants of B. laxiflora with tubers shown at the base. Light microscopy reveals that tuber cells of B. yakushimensis (unstained in C and stained with Sudan Black in D) contain numerous refractile oil droplets. (Scale bar: 50 µM.) (E) EM shows numerous lipid globules (arrows) distributed along the cell wall of bract cells of B. yakushimensis. (Scale bar: 2 µM.) (F) A plastid-like structure in bract cells of B. laxiflora. (Scale bar: 200 nm.) (G) A plastid-like structure in bract cells of B. laxiflora. (Scale bar: 100 nm.)
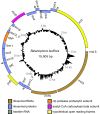
Fig. 2.
Circular map of the B. laxiflora plastome. Genes shown inside and outside the outer circle are transcribed clockwise and counterclockwise, respectively. Triangles mark the two portions, of undetermined extent, of the _trans_-spliced intron in rps12. The four pairs of overlapping genes are marked with inverted “V”s. The histogram shows GC content that is greater than (outside the circle) or less than (inside the circle) the plastome average of 12.2% GC.
Fig. 3.
Evidence for a novel genetic-code change in Balanophora plastomes. (A) Structure of the clpP and rpl2 genes. The approximate location of six codons diagnostic of a code change are given. Exons are represented by boxes and introns by interrupted lines. (B) Partial alignments of the inferred amino acid sequences of CLPP and RPL2. The histograms indicate the percentage of the 18 sequences in the alignment that share the most common amino acid at each position. (C) Pairwise _d_N/_d_S ratios from sliding-window analysis (window size = 90 bp; step size = 30 bp) of the _Balanophora clp_P and _rpl_2 genes (
SI Appendix, Fig. S9
shows the same analysis of three other Balanophora genes). Arrows mark internal TAG codons present in one or both Balanophora plastomes and inferred to encode W; note that, at five of these six positions, most or all non-Balanophora land plants contain TGG (W in the standard genetic code). The positions of these TAG codons in the complete alignments are shown in
SI Appendix, Fig. S4
.
Fig. 4.
Sequence divergence of plastid genes. (A) Phylograms of nonsynonymous (_d_N) and synonymous (_d_S) site divergence for the three indicated genes. (B and C) Levels of _d_N (in black) and _d_S (in light gray) on the branch leading to Balanophora (B) and in pairwise comparisons between the two Balanophora plastomes (C). _d_N/_d_S ratios are given in parentheses.
Comment in
- Evolution: A Plant Plastid Genome that Has Forsaken Guanine and Cytosine.
Smith DR. Smith DR. Curr Biol. 2019 Feb 4;29(3):R99-R101. doi: 10.1016/j.cub.2018.12.025. Curr Biol. 2019. PMID: 30721685
Similar articles
- Plastomes in the holoparasitic family Balanophoraceae: Extremely high AT content, severe gene content reduction, and two independent genetic code changes.
Ceriotti LF, Roulet ME, Sanchez-Puerta MV. Ceriotti LF, et al. Mol Phylogenet Evol. 2021 Sep;162:107208. doi: 10.1016/j.ympev.2021.107208. Epub 2021 May 23. Mol Phylogenet Evol. 2021. PMID: 34029719 - Extreme plastomes in holoparasitic Balanophoraceae are not the norm.
Kim W, Lautenschläger T, Bolin JF, Rees M, Nzuzi A, Zhou R, Wanke S, Jost M. Kim W, et al. BMC Genomics. 2023 Jun 15;24(1):330. doi: 10.1186/s12864-023-09422-1. BMC Genomics. 2023. PMID: 37322447 Free PMC article. - Detecting and Characterizing the Highly Divergent Plastid Genome of the Nonphotosynthetic Parasitic Plant Hydnora visseri (Hydnoraceae).
Naumann J, Der JP, Wafula EK, Jones SS, Wagner ST, Honaas LA, Ralph PE, Bolin JF, Maass E, Neinhuis C, Wanke S, dePamphilis CW. Naumann J, et al. Genome Biol Evol. 2016 Jan 6;8(2):345-63. doi: 10.1093/gbe/evv256. Genome Biol Evol. 2016. PMID: 26739167 Free PMC article. - Invited Review Beyond parasitic convergence: unravelling the evolution of the organellar genomes in holoparasites.
Sanchez-Puerta MV, Ceriotti LF, Gatica-Soria LM, Roulet ME, Garcia LE, Sato HA. Sanchez-Puerta MV, et al. Ann Bot. 2023 Nov 30;132(5):909-928. doi: 10.1093/aob/mcad108. Ann Bot. 2023. PMID: 37503831 Free PMC article. Review. - From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants.
Krause K. Krause K. Curr Genet. 2008 Sep;54(3):111-21. doi: 10.1007/s00294-008-0208-8. Epub 2008 Aug 12. Curr Genet. 2008. PMID: 18696071 Review.
Cited by
- Plastid NDH Pseudogenization and Gene Loss in a Recently Derived Lineage from the Largest Hemiparasitic Plant Genus Pedicularis (Orobanchaceae).
Li X, Yang JB, Wang H, Song Y, Corlett RT, Yao X, Li DZ, Yu WB. Li X, et al. Plant Cell Physiol. 2021 Oct 11;62(6):971-984. doi: 10.1093/pcp/pcab074. Plant Cell Physiol. 2021. PMID: 34046678 Free PMC article. - Evolution of a Record-Setting AT-Rich Genome: Indel Mutation, Recombination, and Substitution Bias.
Nguyen DT, Wu B, Xiao S, Hao W. Nguyen DT, et al. Genome Biol Evol. 2020 Dec 6;12(12):2344-2354. doi: 10.1093/gbe/evaa202. Genome Biol Evol. 2020. PMID: 32986811 Free PMC article. - What we know so far and what we can expect next: A molecular investigation of plant parasitism.
Ishida JK, Costa EC. Ishida JK, et al. Genet Mol Biol. 2024 Sep 30;47Suppl 1(Suppl 1):e20240051. doi: 10.1590/1678-4685-GMB-2024-0051. eCollection 2024. Genet Mol Biol. 2024. PMID: 39348487 Free PMC article. - Cytonuclear coevolution in a holoparasitic plant with highly disparate organellar genomes.
Ceriotti LF, Gatica-Soria L, Sanchez-Puerta MV. Ceriotti LF, et al. Plant Mol Biol. 2022 Aug;109(6):673-688. doi: 10.1007/s11103-022-01266-9. Epub 2022 Mar 31. Plant Mol Biol. 2022. PMID: 35359176 - Systematics and Plastome Evolution in Schizaeaceae.
Ke BF, Wang GJ, Labiak PH, Rouhan G, Chen CW, Shepherd LD, Ohlsen DJ, Renner MAM, Karol KG, Li FW, Kuo LY. Ke BF, et al. Front Plant Sci. 2022 Jul 13;13:885501. doi: 10.3389/fpls.2022.885501. eCollection 2022. Front Plant Sci. 2022. PMID: 35909781 Free PMC article.
References
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. - PubMed
- Leake JR. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994;127:171–216. - PubMed
- Krause K. From chloroplasts to “cryptic” plastids: Evolution of plastid genomes in parasitic plants. Curr Genet. 2008;54:111–121. - PubMed
- Barrett CF, Davis JI. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 2012;99:1513–1523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources