Plastome-Wide Rearrangements and Gene Losses in Carnivorous Droseraceae - PubMed (original) (raw)
Plastome-Wide Rearrangements and Gene Losses in Carnivorous Droseraceae
Paul G Nevill et al. Genome Biol Evol. 2019.
Abstract
The plastid genomes of four related carnivorous plants (Drosera regia, Drosera erythrorhiza, Aldrovanda vesiculosa, and Dionaea muscipula) were sequenced to examine changes potentially induced by the transition to carnivory. The plastid genomes of the Droseraceae show multiple rearrangements, gene losses, and large expansions or contractions of the inverted repeat. All the ndh genes are lost or nonfunctional, as well as in some of the species, clpP1, ycf1, ycf2 and some tRNA genes. Uniquely, among land plants, the trnK gene has no intron. Carnivory in the Droseraceae coincides with changes in plastid gene content similar to those induced by parasitism and mycoheterotrophy, suggesting parallel changes in chloroplast function due to the similar switch from autotrophy to (mixo-) heterotrophy. A molecular phylogeny of the taxa based on all shared plastid genes indicates that the "snap-traps" of Aldrovanda and Dionaea have a common origin.
Keywords: Droseraceae; carnivorous plants; chloroplast genome; gene loss; intron loss.
© The Author(s) 2018. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
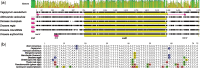
Fig. 1.
—Physical maps of the plastid genomes of Droseraceae compared with that of Fagopyrum esculentum. The genomes are drawn to scale. Different gene classes and the inverted repeats are indicated by colored bands. Gray lines connecting adjacent genomes indicate synteny; each line represents an indel-free sequence alignment of >40 bp in length and with >50% matching bases. The genomes have been arranged to display synteny conservation as much as possible, that is, the most divergent genomes have been placed at the top and bottom.
Fig. 2.
—Gene content in Droseraceae plastid genomes and in Fagopyrum esculentum. Genes present and apparently functional are shaded in black, doubtfully functional genes in mid-gray, almost certainly nonfunctional remnants in pale gray and missing genes in white.
Fig. 3.
—Mapping of Drosera erythrorhiza reads to the Drosera regia genome across the inverted repeat-single copy region. Reads were mapped using bowtie2 (Langmead and Salzberg 2012) with default parameters and coverage was visualized using Geneious 9.1.5 (
, last accessed February 3, 2019, Kearse et al. 2012). Most genes in this region show extensive coverage (∼1,000×), but coverage is extremely low over the last and first exons of trnI and trnA, respectively, almost all of ycf1, and rpl32. The lack of crossmapping of raw reads to Drosera regia confirms that these sequences are missing from the Drosera erythrorhiza genome.
Fig. 4.
—(a) Alignment of the predicted accD protein sequences from the Droseraceae species compared with that of Fagopyrum esculentum, shaded to indicate conservation. Black indicates amino acids identical to those in the F. esculentum sequence. Sequences are numbered as follows: 1, F. esculentum; 2, Aldrovanda vesiculosa; 3, Dionaea muscipula; 4, Drosera regia; 5, Drosera erythrorhiza; and 6, Drosera rotundifolia. The sequences from D. muscipula, Dr. erythrorhiza, and Dr. rotundifolia are truncated at the N-terminus, contain large indels, and in the case of D. muscipula, a long C-terminal extension due to mutation of the usual stop codon_._ The functionality of these sequences is doubtful. (b) Alignment of the trnG-UCC gene from the Droseraceae species compared with that of F. esculentum, shaded to indicate conservation. Black indicates nucleotides identical to those in the F. esculentum sequence. The positions of the tRNA exons are indicated by magenta arrows. Sequences are numbered as in (a). Alignment and visualization was done with Geneious 9.1.5 (
, Kearse et al. 2012). (c) Cloverleaf representation of the trnG-UCC sequence from F. esculentum, indicating nucleotide changes in the Droseraceae (magenta for D. muscipula, orange for Dr. rotundifolia, and red for Dr. sera erythrorhiza). Many of these nucleotide changes would be expected to severely impact the function of the tRNA, most obviously the alteration in the anticodon in Dr. erythrorhiza.
Fig. 5.
—(a) Alignment of the trnK-matK-psbA region of the Droseraceae genomes in comparison to the homologous region from Fagopyrum esculentum. The matK coding sequence is indicated in yellow, tRNA genes in magenta (trnK in all cases except Drosera erythrorhiza, where the tRNA gene shown is trnV-GAC; trnK is found elsewhere in the genome). Nucleotide identity is indicated by the plot about the alignment (green indicates 100% identity, yellow indicates 30–80%, and red indicates below 30%). Alignment and visualization was done with Geneious 9.1.5 (
, Kearse et al. 2012). (b) Alignment of trnK sequences from diverse plastids. The sequences were selected as representatives from an alignment of 1654 trnK and tRNA-Lys sequences from GenBank. Dicot and monocot consensus sequences represent hundreds of identical sequences from these clades. The other sequences shown are from specific accessions: Fagopyrum esculentum (NC_010776), Stangeria eriopus (JX416858.1), Marchantia polymorpha (M20959.1), Drosera regia (this work), Grateloupia taiwanensis (KC894740.1), Thorea hispida (KX284714.1), Pseudomuriella schumacherensis (KT199256.1), and Cyclospora cayetanensis (KX273388.1). The trnK genes in land plants (except the Droseraceae) are split by an intron between positions 37–38, so the sequences shown are the exon fusions. Those in algae (except charophytes) are not split.
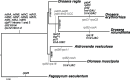
Fig. 6.
—Phylogenetic tree based on Droseraceae plastid genomes. Fifty-eight protein-coding sequences shared by all genomes in this study were aligned gene-by-gene and then the alignments were concatenated. The most probable tree topology obtained by MrBayes, RAxML, and IQ-TREE is shown with the support values (posterior probabilities or bootstrap support) shown at the nodes. The branch lengths calculated by each program were similar. Putative loss of gene function, intron losses, and breaks in cotranscription units (in gray text) are indicated against the branches where they occurred.
Similar articles
- Genomes of the Venus Flytrap and Close Relatives Unveil the Roots of Plant Carnivory.
Palfalvi G, Hackl T, Terhoeven N, Shibata TF, Nishiyama T, Ankenbrand M, Becker D, Förster F, Freund M, Iosip A, Kreuzer I, Saul F, Kamida C, Fukushima K, Shigenobu S, Tamada Y, Adamec L, Hoshi Y, Ueda K, Winkelmann T, Fuchs J, Schubert I, Schwacke R, Al-Rasheid K, Schultz J, Hasebe M, Hedrich R. Palfalvi G, et al. Curr Biol. 2020 Jun 22;30(12):2312-2320.e5. doi: 10.1016/j.cub.2020.04.051. Epub 2020 May 14. Curr Biol. 2020. PMID: 32413308 Free PMC article. - Distinctive plastome evolution in carnivorous angiosperms.
Fu CN, Wicke S, Zhu AD, Li DZ, Gao LM. Fu CN, et al. BMC Plant Biol. 2023 Dec 20;23(1):660. doi: 10.1186/s12870-023-04682-1. BMC Plant Biol. 2023. PMID: 38124058 Free PMC article. - Evolution of genome size and genomic GC content in carnivorous holokinetics (Droseraceae).
Veleba A, Šmarda P, Zedek F, Horová L, Šmerda J, Bureš P. Veleba A, et al. Ann Bot. 2017 Feb;119(3):409-416. doi: 10.1093/aob/mcw229. Epub 2016 Dec 26. Ann Bot. 2017. PMID: 28025291 Free PMC article. - Trap diversity and evolution in the family Droseraceae.
Poppinga S, Hartmeyer SR, Masselter T, Hartmeyer I, Speck T. Poppinga S, et al. Plant Signal Behav. 2013 Jul;8(7):e24685. doi: 10.4161/psb.24685. Epub 2013 Apr 18. Plant Signal Behav. 2013. PMID: 23603942 Free PMC article. Review. - On the Origin of Carnivory: Molecular Physiology and Evolution of Plants on an Animal Diet.
Hedrich R, Fukushima K. Hedrich R, et al. Annu Rev Plant Biol. 2021 Jun 17;72:133-153. doi: 10.1146/annurev-arplant-080620-010429. Epub 2021 Jan 12. Annu Rev Plant Biol. 2021. PMID: 33434053 Review.
Cited by
- Towards the plastome evolution and phylogeny of Cycas L. (Cycadaceae): molecular-morphology discordance and gene tree space analysis.
Liu J, Lindstrom AJ, Gong X. Liu J, et al. BMC Plant Biol. 2022 Mar 15;22(1):116. doi: 10.1186/s12870-022-03491-2. BMC Plant Biol. 2022. PMID: 35291941 Free PMC article. - Large scale genome skimming from herbarium material for accurate plant identification and phylogenomics.
Nevill PG, Zhong X, Tonti-Filippini J, Byrne M, Hislop M, Thiele K, van Leeuwen S, Boykin LM, Small I. Nevill PG, et al. Plant Methods. 2020 Jan 4;16:1. doi: 10.1186/s13007-019-0534-5. eCollection 2020. Plant Methods. 2020. PMID: 31911810 Free PMC article. - Intraspecific Variation within the Utricularia amethystina Species Morphotypes Based on Chloroplast Genomes.
Silva SR, Pinheiro DG, Penha HA, Płachno BJ, Michael TP, Meer EJ, Miranda VFO, Varani AM. Silva SR, et al. Int J Mol Sci. 2019 Dec 5;20(24):6130. doi: 10.3390/ijms20246130. Int J Mol Sci. 2019. PMID: 31817365 Free PMC article. - Phylotranscriptomic Analyses of Mycoheterotrophic Monocots Show a Continuum of Convergent Evolutionary Changes in Expressed Nuclear Genes From Three Independent Nonphotosynthetic Lineages.
Timilsena PR, Barrett CF, Piñeyro-Nelson A, Wafula EK, Ayyampalayam S, McNeal JR, Yukawa T, Givnish TJ, Graham SW, Pires JC, Davis JI, Ané C, Stevenson DW, Leebens-Mack J, Martínez-Salas E, Álvarez-Buylla ER, dePamphilis CW. Timilsena PR, et al. Genome Biol Evol. 2023 Jan 4;15(1):evac183. doi: 10.1093/gbe/evac183. Genome Biol Evol. 2023. PMID: 36582124 Free PMC article. - Comparative plastomics of Amaryllidaceae: inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq.
Könyves K, Bilsborrow J, Christodoulou MD, Culham A, David J. Könyves K, et al. PeerJ. 2021 Nov 12;9:e12400. doi: 10.7717/peerj.12400. eCollection 2021. PeerJ. 2021. PMID: 34824912 Free PMC article.
References
- Adamec L. 1997. Mineral nutrition of carnivorous plants: a review. Botanical Rev. 63(3):273–299.
- Adlassnig W, et al. 2012. Endocytotic uptake of nutrients in carnivorous plants. Plant J. 71(2):303–313. - PubMed
- Albert VA, Williams SE, Chase MW.. 1992. Carnivorous plants: phylogeny and structural evolution. Science 257(5076):1491–1495. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources