Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use - PubMed (original) (raw)
Meta-Analysis
. 2019 Feb;51(2):237-244.
doi: 10.1038/s41588-018-0307-5. Epub 2019 Jan 14.
Yu Jiang 2 3, Robbee Wedow 4 5 6, Yue Li 7 8, David M Brazel 4 9 10, Fang Chen 2 3, Gargi Datta 1, Jose Davila-Velderrain 7 8, Daniel McGuire 2 3, Chao Tian 11, Xiaowei Zhan 12 13; 23andMe Research Team; HUNT All-In Psychiatry; Hélène Choquet 14, Anna R Docherty 15 16, Jessica D Faul 17, Johanna R Foerster 18, Lars G Fritsche 18, Maiken Elvestad Gabrielsen 19, Scott D Gordon 20, Jeffrey Haessler 21, Jouke-Jan Hottenga 22, Hongyan Huang 23 24, Seon-Kyeong Jang 1, Philip R Jansen 25 26, Yueh Ling 2 9, Reedik Mägi 27, Nana Matoba 28, George McMahon 29, Antonella Mulas 30, Valeria Orrù 30, Teemu Palviainen 31, Anita Pandit 18, Gunnar W Reginsson 32, Anne Heidi Skogholt 19, Jennifer A Smith 17 33, Amy E Taylor 29, Constance Turman 23 24, Gonneke Willemsen 22, Hannah Young 1, Kendra A Young 34, Gregory J M Zajac 18, Wei Zhao 33, Wei Zhou 35, Gyda Bjornsdottir 32, Jason D Boardman 4 5 6, Michael Boehnke 18, Dorret I Boomsma 22, Chu Chen 21, Francesco Cucca 30, Gareth E Davies 36, Charles B Eaton 37, Marissa A Ehringer 4 38, Tõnu Esko 8 27, Edoardo Fiorillo 30, Nathan A Gillespie 15 20, Daniel F Gudbjartsson 32 39, Toomas Haller 27, Kathleen Mullan Harris 40 41, Andrew C Heath 42, John K Hewitt 4 43, Ian B Hickie 44, John E Hokanson 34, Christian J Hopfer 4 45, David J Hunter 23 24 46, William G Iacono 1, Eric O Johnson 47, Yoichiro Kamatani 28, Sharon L R Kardia 33, Matthew C Keller 4 43, Manolis Kellis 7 8, Charles Kooperberg 21, Peter Kraft 23 24 48, Kenneth S Krauter 4 9, Markku Laakso 49 50, Penelope A Lind 51, Anu Loukola 31, Sharon M Lutz 52, Pamela A F Madden 42, Nicholas G Martin 20, Matt McGue 1, Matthew B McQueen 4 38, Sarah E Medland 51, Andres Metspalu 27, Karen L Mohlke 53, Jonas B Nielsen 54, Yukinori Okada 28 55, Ulrike Peters 21 56, Tinca J C Polderman 25, Danielle Posthuma 25 57, Alexander P Reiner 21 56, John P Rice 58, Eric Rimm 24 59, Richard J Rose 60, Valgerdur Runarsdottir 61, Michael C Stallings 4 43, Alena Stančáková 49, Hreinn Stefansson 32, Khanh K Thai 14, Hilary A Tindle 62, Thorarinn Tyrfingsson 61, Tamara L Wall 63, David R Weir 17, Constance Weisner 14, John B Whitfield 20, Bendik Slagsvold Winsvold 64, Jie Yin 14, Luisa Zuccolo 29 65, Laura J Bierut 58, Kristian Hveem 19 66 67, James J Lee 1, Marcus R Munafò 65 68, Nancy L Saccone 69, Cristen J Willer 35 54 70, Marilyn C Cornelis 71, Sean P David 72, David A Hinds 11, Eric Jorgenson 14, Jaakko Kaprio 31 73, Jerry A Stitzel 4 38, Kari Stefansson 32 74, Thorgeir E Thorgeirsson 32, Gonçalo Abecasis 18, Dajiang J Liu 75 76, Scott Vrieze 77
Collaborators, Affiliations
- PMID: 30643251
- PMCID: PMC6358542
- DOI: 10.1038/s41588-018-0307-5
Meta-Analysis
Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use
Mengzhen Liu et al. Nat Genet. 2019 Feb.
Abstract
Tobacco and alcohol use are leading causes of mortality that influence risk for many complex diseases and disorders1. They are heritable2,3 and etiologically related4,5 behaviors that have been resistant to gene discovery efforts6-11. In sample sizes up to 1.2 million individuals, we discovered 566 genetic variants in 406 loci associated with multiple stages of tobacco use (initiation, cessation, and heaviness) as well as alcohol use, with 150 loci evidencing pleiotropic association. Smoking phenotypes were positively genetically correlated with many health conditions, whereas alcohol use was negatively correlated with these conditions, such that increased genetic risk for alcohol use is associated with lower disease risk. We report evidence for the involvement of many systems in tobacco and alcohol use, including genes involved in nicotinic, dopaminergic, and glutamatergic neurotransmission. The results provide a solid starting point to evaluate the effects of these loci in model organisms and more precise substance use measures.
Conflict of interest statement
COMPETING INTERESTS STATEMENT: Laura J. Bierut and the spouse of Nancy L. Saccone are listed as inventors on Issued U.S. Patent 8,080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Sean David is a scientific advisor to BaseHealth, Inc. Gyda Bjornsdottir, Daniel F. Gudbjartsson, Gunnar W. Reginsson, Hreinn Stefansson, Kari Stefansson, and Thorgeir E. Thorgeirsson are employees of deCODE Genetics/AMGEN, Inc. Chao Tian and David Hinds are employees of 23andMe, Inc.
Figures
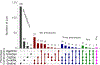
Figure 1.. Genetic correlations between substance use phenotypes and phenotypes from other large genome-wide association studies.
Genetic correlations between each of the phenotypes are shown in the first 5 rows, with heritability estimates displayed down the diagonal. All genetic correlations and heritability estimates were calculated using LD Score Regression. Blue shading represents negative genetic correlations, and red shading represents positive correlations, with increasing color intensity reflecting increasing strength of a correlation. A single asterisk reflects significant genetic correlations at the p<.05 level. Double asterisks reflect significant genetic correlations at the Bonferroni-correction p<.000278 level (corrected for 180 independent tests). Note that SmkCes was oriented such that higher scores reflected current smoking, and for AgeSmk lower scores reflect earlier ages of initiation, both of which are typically associated with negative outcomes. AgeSmk=Age of Initiation of Smoking; CigDay=Cigarettes per Day; SmkInit=Smoking Initiation; SmkCes=Smoking Cessation; DrnkWk=Drinks per Week.
Figure 2.. Pleiotropy.
Depicted here are results from the multivariate analysis of pleiotropy. For each locus, the method returns the best fitting solution of which phenotypes were associated with that locus. All loci with one or more associated phenotypes are shown here. For example, every locus associated with AgeSmk was found to be pleiotropic for other phenotypes (green, blue, red, purple, and fuchsia bars), and no locus showed association with only AgeSmk (no dark grey bar for AgeSmk). When sample sizes are unequal across phenotypes, the method also improves power for those phenotypes with smaller samples. The total number of loci associated with each trait (whether pleiotropic or not) from these analyses was 40 (AgeSmk), 48 (SmkCes), 72 (CigDay), 111 (DrnkWk), and 278 (SmkInit). Full information is in Supplementary Table 11.
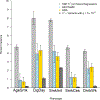
Figure 3.. Heritability and polygenic prediction.
The light gray bars reflect SNP heritability, estimated with LD Score Regression. The light blue and gold bars reflect the predictive power of polygenic risk scores in Add Health and the Health and Retirement Study (HRS), respectively. Despite the 41-year generational gap between participants from these two studies, and major tobacco-related policy changes during that time, the polygenic scores are similarly predictive in both samples. Error bars are 95% confidence intervals estimated with 1000 bootstrapped repetitions. Dark gray bars represent the total phenotypic variance explained by only genome-wide significant SNPs. _H_2=heritability.
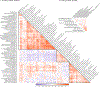
Figure 4.. Correlations among exemplary DEPICT gene sets.
There were 68 clusters available for Smoking initiation and 10 for Drinks Per Week (CigDay, AgeSmk, and SmkCes did not have > 1 exemplary sets.) Blue shading represents positive correlations, and red shading represents negative correlations, with increasing color intensity reflecting increasing strength of a correlation. Cluster names are truncated for space, with a full list of all names in Supplementary Table 18. The number after each name is the number of gene sets in each cluster. The matrix naturally falls into three blue superclusters along the diagonal. The largest supercluster contains primarily gene sets related to neurotransmitter receptors, ion channels (sodium, potassium, calcium), learning/memory, and other aspects of CNS function. The middle supercluster includes gene sets defined by regulation of transcription and translation, including RNA binding and transcription factor activity. The final supercluster is composed primarily of gene sets related to development of the nervous system.
References
- Ezzati M et al. Selected major risk factors and global and regional burden of disease. Lancet 360, 1347–1360 (2002). - PubMed
- Polderman TJ et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet (2015). - PubMed
- Kendler KS, Prescott CA, Myers J & Neale MC The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Archives of General Psychiatry 60, 929–937 (2003). - PubMed
METHODS ONLY REFERENCES
- Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38, 904–909 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
- R01 DK093757/DK/NIDDK NIH HHS/United States
- R01 HL089897/HL/NHLBI NIH HHS/United States
- P01 HD031921/HD/NICHD NIH HHS/United States
- K01 HL125858/HL/NHLBI NIH HHS/United States
- U01 AG009740/AG/NIA NIH HHS/United States
- R03 OD032630/OD/NIH HHS/United States
- R01 GM126479/GM/NIGMS NIH HHS/United States
- R03 HD097630/HD/NICHD NIH HHS/United States
- R01 DA042755/DA/NIDA NIH HHS/United States
- G0902144/MRC_/Medical Research Council/United Kingdom
- P2C HD066613/HD/NICHD NIH HHS/United States
- T32 DK091317/DK/NIDDK NIH HHS/United States
- K01 MH109765/MH/NIMH NIH HHS/United States
- R01 MH123619/MH/NIMH NIH HHS/United States
- R03 DC013373/DC/NIDCD NIH HHS/United States
- R21 DA040177/DA/NIDA NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- MC_UU_00011/7/MRC_/Medical Research Council/United Kingdom
- R01 DK072193/DK/NIDDK NIH HHS/United States
- MR/K023195/1/MRC_/Medical Research Council/United Kingdom
- T32 HG000040/HG/NHGRI NIH HHS/United States
- R01 HL119443/HL/NHLBI NIH HHS/United States
- R01 DK062370/DK/NIDDK NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- R01 DA034076/DA/NIDA NIH HHS/United States
- R01 HG008983/HG/NHGRI NIH HHS/United States
- R01 DA036583/DA/NIDA NIH HHS/United States
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- U01 DK062370/DK/NIDDK NIH HHS/United States
- R21 AA021223/AA/NIAAA NIH HHS/United States
- R01 DA037904/DA/NIDA NIH HHS/United States
- U01 DA046413/DA/NIDA NIH HHS/United States
- R01 AA023974/AA/NIAAA NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- S10 OD020069/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases