DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays - PubMed (original) (raw)
DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays
Amrit Singh et al. Bioinformatics. 2019.
Abstract
Motivation: In the continuously expanding omics era, novel computational and statistical strategies are needed for data integration and identification of biomarkers and molecular signatures. We present Data Integration Analysis for Biomarker discovery using Latent cOmponents (DIABLO), a multi-omics integrative method that seeks for common information across different data types through the selection of a subset of molecular features, while discriminating between multiple phenotypic groups.
Results: Using simulations and benchmark multi-omics studies, we show that DIABLO identifies features with superior biological relevance compared with existing unsupervised integrative methods, while achieving predictive performance comparable to state-of-the-art supervised approaches. DIABLO is versatile, allowing for modular-based analyses and cross-over study designs. In two case studies, DIABLO identified both known and novel multi-omics biomarkers consisting of mRNAs, miRNAs, CpGs, proteins and metabolites.
Availability and implementation: DIABLO is implemented in the mixOmics R Bioconductor package with functions for parameters' choice and visualization to assist in the interpretation of the integrative analyses, along with tutorials on http://mixomics.org and in our Bioconductor vignette.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
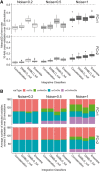
Fig. 1.
Simulation study. (A) Classification error rates (10-fold CV averaged over 20 simulations) for different FCs between groups and varying level of noise (SD). Dashed line indicates a random performance (error rate = 50%). (B) Types of variables selected by the different classification methods amongst the 180 variables selected for each classification method
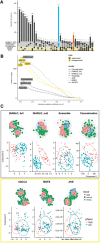
Fig. 2.
Benchmark for colon cancer. (A) Number of selected features overlapping between supervised and unsupervised methods. (B) Number of correlated variables in the biomarker panels for various Pearson correlation cutoffs. (C) Top: network modularity of each multi-omics biomarker panel. Gray circles depict modules based on the edge betweenness index from the igraph R-library. Bottom: consensus component plots depicting the separation of subjects in the high and low survival groups. Similar patterns were observed for kidney, gbm and lung cancer datasets, see Supplementary Figures S5–S10
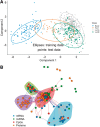
Fig. 3.
A multi-omics biomarker panel predictive of breast cancer subtypes. (A) DIABLO consensus component plot based on the identified multi-omics biomarker panel: test samples are overlaid with 95% confidence ellipses calculated from the training data. (B) Network visualization of the biomarker panel highlighting correlated variables (absolute Pearson’s correlation >0.4) and four communities based on the edge betweenness index
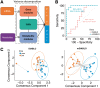
Fig. 4.
Asthma study: cross-over design and module-based analysis. (A) DIABLO design includes module-based decomposition to discriminate pre- and post-allergen challenge samples. (B) Receiver operating characteristic curves comparing standard DIABLO and multilevel DIABLO for repeated measures (mDIABLO) using leave-one-out CV. (C) Component plots of the pre- and post-challenge samples (DIABLO and mDIABLO)
Similar articles
- mixOmics: An R package for 'omics feature selection and multiple data integration.
Rohart F, Gautier B, Singh A, Lê Cao KA. Rohart F, et al. PLoS Comput Biol. 2017 Nov 3;13(11):e1005752. doi: 10.1371/journal.pcbi.1005752. eCollection 2017 Nov. PLoS Comput Biol. 2017. PMID: 29099853 Free PMC article. - timeOmics: an R package for longitudinal multi-omics data integration.
Bodein A, Scott-Boyer MP, Perin O, Lê Cao KA, Droit A. Bodein A, et al. Bioinformatics. 2022 Jan 3;38(2):577-579. doi: 10.1093/bioinformatics/btab664. Bioinformatics. 2022. PMID: 34554215 - Machine learning combining multi-omics data and network algorithms identifies adrenocortical carcinoma prognostic biomarkers.
Martin-Hernandez R, Espeso-Gil S, Domingo C, Latorre P, Hervas S, Hernandez Mora JR, Kotelnikova E. Martin-Hernandez R, et al. Front Mol Biosci. 2023 Nov 6;10:1258902. doi: 10.3389/fmolb.2023.1258902. eCollection 2023. Front Mol Biosci. 2023. PMID: 38028548 Free PMC article. - Identifying biomarkers for predicting successful embryo implantation: applying single to multi-OMICs to improve reproductive outcomes.
Hernández-Vargas P, Muñoz M, Domínguez F. Hernández-Vargas P, et al. Hum Reprod Update. 2020 Feb 28;26(2):264-301. doi: 10.1093/humupd/dmz042. Hum Reprod Update. 2020. PMID: 32096829 Review. - A Detailed Catalogue of Multi-Omics Methodologies for Identification of Putative Biomarkers and Causal Molecular Networks in Translational Cancer Research.
Vlachavas EI, Bohn J, Ückert F, Nürnberg S. Vlachavas EI, et al. Int J Mol Sci. 2021 Mar 10;22(6):2822. doi: 10.3390/ijms22062822. Int J Mol Sci. 2021. PMID: 33802234 Free PMC article. Review.
Cited by
- Integrative microbiome and metabolome profiles reveal the impacts of periodontitis via oral-gut axis in first-trimester pregnant women.
Cheng T, Wen P, Yu R, Zhang F, Li H, Xu X, Zhao D, Liu F, Su W, Zheng Z, Yang H, Yao J, Jin L. Cheng T, et al. J Transl Med. 2024 Sep 3;22(1):819. doi: 10.1186/s12967-024-05579-9. J Transl Med. 2024. PMID: 39227984 Free PMC article. - Molecular Signatures of Idiopathic Pulmonary Fibrosis.
Konigsberg IR, Borie R, Walts AD, Cardwell J, Rojas M, Metzger F, Hauck SM, Fingerlin TE, Yang IV, Schwartz DA. Konigsberg IR, et al. Am J Respir Cell Mol Biol. 2021 Oct;65(4):430-441. doi: 10.1165/rcmb.2020-0546OC. Am J Respir Cell Mol Biol. 2021. PMID: 34038697 Free PMC article. - Comparative analysis of integrative classification methods for multi-omics data.
Novoloaca A, Broc C, Beloeil L, Yu WH, Becker J. Novoloaca A, et al. Brief Bioinform. 2024 May 23;25(4):bbae331. doi: 10.1093/bib/bbae331. Brief Bioinform. 2024. PMID: 38985929 Free PMC article. - Network-based integration of multi-omics data for clinical outcome prediction in neuroblastoma.
Wang C, Lue W, Kaalia R, Kumar P, Rajapakse JC. Wang C, et al. Sci Rep. 2022 Sep 14;12(1):15425. doi: 10.1038/s41598-022-19019-5. Sci Rep. 2022. PMID: 36104347 Free PMC article. - Microbe-Immune Crosstalk: Evidence That T Cells Influence the Development of the Brain Metabolome.
Caspani G, Green M, Swann JR, Foster JA. Caspani G, et al. Int J Mol Sci. 2022 Mar 17;23(6):3259. doi: 10.3390/ijms23063259. Int J Mol Sci. 2022. PMID: 35328680 Free PMC article.
References
- Aben N., et al. (2016) Tandem: a two-stage approach to maximize interpretability of drug response models based on multiple molecular data types. Bioinformatics, 32, i413–i420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources