A subtelomeric region affects telomerase-negative replicative senescence in Saccharomyces cerevisiae - PubMed (original) (raw)
A subtelomeric region affects telomerase-negative replicative senescence in Saccharomyces cerevisiae
Pascale Jolivet et al. Sci Rep. 2019.
Abstract
In eukaryotes, telomeres determine cell proliferation potential by triggering replicative senescence in the absence of telomerase. In Saccharomyces cerevisiae, senescence is mainly dictated by the first telomere that reaches a critically short length, activating a DNA-damage-like response. How the corresponding signaling is modulated by the telomeric structure and context is largely unknown. Here we investigated how subtelomeric elements of the shortest telomere in a telomerase-negative cell influence the onset of senescence. We found that a 15 kb truncation of the 7L subtelomere widely used in studies of telomere biology affects cell growth when combined with telomerase inactivation. This effect is likely not explained by (i) elimination of sequence homology at chromosome ends that would compromise homology-directed DNA repair mechanisms; (ii) elimination of the conserved subtelomeric X-element; (iii) elimination of a gene that would become essential in the absence of telomerase; and (iv) heterochromatinization of inner genes, causing the silencing of an essential gene in replicative senescent cells. This works contributes to better delineate subtelomere functions and their impact on telomere biology.
Conflict of interest statement
The authors declare no competing interests.
Figures
Figure 1
Experimental system to shorten a single telomere in the cell. The chromosome end containing the 7L telomere (a) is modified in two ways. In control (CTL) cells (b), the last 15 kb of the telomere end are replaced by a construct in which a URA3 marker is flanked by two Flippase Recognition Target (FRT) sites and followed by a wild-type length telomeric tract. (c) In cells able to generate a very short telomere (VST), the URA3 marker is followed by extratelomeric repeats that inhibit the action of telomerase on telomeric repeats in cis. This results in a short terminal telomeric tract. (b,c) Upon telomerase removal and induction of flippase (encoded by FLP1), sequences between the FRT sites are excised in a circle that is diluted out upon successive cell divisions. Remaining telomeres are of wild-type length (b) or very short (c). (d) The wild type 6R chromosome end is modified to generate 6R-CTL (e) and 6R-VST (f) in a manner similar to 7L (a–c). Chromosome maps and annotations are from the Saccharomyces Genome Database and Centre for Genetic Architecture of Complex Traits website (
https://www.le.ac.uk/colleges/medbiopsych/research/gact/resources/yeast-telomeres
) using the sequence of strain S288C as reference. Red and orange colors correspond to genes encoding minimally characterized proteins or dubious/putative proteins, respectively. Note that subtelomeric sequences of strains W303 – used in this work - and S288C – used as reference - 7L diverge in a region spanning Ty5 to YGL262W.
Figure 2
Effect of the subtelomeric region on replicative senescence. 16 telomerase-negative individual spores carrying the telomere 7L-CTL (blue), 7L-VST (red), 6R-CTL (black) or 6R-VST (purple) (see Fig. 1b,c, e,f) were germinated for two days on selective media. Colonies grown on selective plates for 2 days were then resuspended to equal concentrations and 10-fold dilutions were spotted on solid media, grown at 30 °C for 2 days (passage 1). This procedure was repeated twice (passage 2 and 3). (a) Cells from passage 1 were used to prepare DNA and telomere length measurements were performed by telomere-PCR using specific primers amplifying either the 7L or the 6R-derived telomeres. Median telomere length is shown. Error bars correspond to SD. Adjusted p-values were obtained by the Wilcoxon rank-sum test with a false discovery rate correction **p < 0.01 (n = 14, 14, 16 and 9, respectively). Plates were scanned at high resolution (b) and analyzed to obtain a numerical value for each serial dilution set that is related to the intensity of the spots (c). Adjusted p-values were obtained by the Wilcoxon rank-sum test with a false discovery rate correction *p-value < 0.05, **p-value < 0.01 and ***p-value < 0,001. n = 16 for 7L-CTL, 6R-CTL and 6R-VST, n = 15 for 7L-VST. See Supplementary Table 3 for detailed p-values.
Figure 3
The homologous recombination machinery counteracts senescence both in the presence and absence of subtelomeric elements at the VST. (a) The absence of Rad51 accelerates replicative senescence in the presence or absence of subtelomeric elements at the shortest telomere. Quantitative analysis of serial spot assays of senescence for cells carrying 7L-VST or 6R-VST as in Fig. 2c. Adjusted p-values were obtained by the Wilcoxon rank-sum test with a false discovery rate correction *p-value < 0.05, **p-value < 0.01 and ***p-value < 0,001. n = 7, 6, 5, 5 for each strain set respectively. See Supplementary Table 3 for detailed p-values. (b) Rad51 and Rad52 preferentially associate with short telomeres in the presence and absence of subtelomere. For each indicated strain, 7L-CTL, 7L-VST, 6R-CTL or 6R-VST, a mixed population of hundreds of independent telomerase-negative clones was grown for ~30 population doublings after sporulation. Chromatin was immunoprecipitated using primary antibodies against either Rad52 (left panel) or Rad51 (right panel). The association of each protein to 7L, 6R or Y’ telomeres or to the ARO1 locus was quantified by qPCR and the fold increase of telomere enrichment over ARO1 is represented. Two biologically independent experiments are shown.
Figure 4
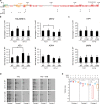
Subtelomeres affect senescence independently of the X-element. (a) Scheme of the deletion of the15 kb, the terminal 8 kb, and the internal 8–14 kb of the sub-telomere 7L. Annotations are as in Fig. 1a. (b,d,f) Spores of the indicated genotype were spotted in parallel for three serial passages and analyzed as in Fig. 2b,c (c,e,g). Adjusted P-values were obtained by the Wilcoxon rank-sum test with a false discovery rate correction *p-value < 0.05, **p-value < 0.01 and ***p-value < 0,001. n = 8 for each strain set. See Supplementary Table 3 for detailed p-values.
Figure 5
Chromatin is likely unchanged in strains lacking terminal 15 kb of 7L. (a) Genetic elements overexpressed upon methyl methanesulfonate (MMS) treatment, hydroxyurea (HU) treatment, or in tlc1- Δ are indicated with “+” signs, according to,. (b,c) mRNA levels of indicated coding sequences in indicated strains were assessed by qPCR, normalized by ACT1 mRNA and by corresponding levels in TLC1 7L-WT strains. Error bars correspond to SD. *p-value < 0.05 obtained with one-tailed Student T-test. n ≥ 3. (d) Spores of the indicated genotype were spotted in parallel for three serial passages on YPD or YPD complemented with 5 mM Nicotinamide (NAM) and analyzed as in Fig. 2b,c (e). Adjusted P-values were obtained by the Wilcoxon rank-sum test with a false discovery rate correction *p-value < 0.05 and **p-value < 0.01. n = 8 for each strain set. See Supplementary Table 3 for detailed p-values.
Similar articles
- Inhibition of the alternative lengthening of telomeres pathway by subtelomeric sequences in Saccharomyces cerevisiae.
Grandin N, Gallego ME, White CI, Charbonneau M. Grandin N, et al. DNA Repair (Amst). 2020 Dec;96:102996. doi: 10.1016/j.dnarep.2020.102996. Epub 2020 Oct 19. DNA Repair (Amst). 2020. PMID: 33126043 - The length of the shortest telomere as the major determinant of the onset of replicative senescence.
Xu Z, Duc KD, Holcman D, Teixeira MT. Xu Z, et al. Genetics. 2013 Aug;194(4):847-57. doi: 10.1534/genetics.113.152322. Epub 2013 Jun 3. Genetics. 2013. PMID: 23733785 Free PMC article. - Tel1/ATM Signaling to the Checkpoint Contributes to Replicative Senescence in the Absence of Telomerase.
Menin L, Colombo CV, Maestrini G, Longhese MP, Clerici M. Menin L, et al. Genetics. 2019 Oct;213(2):411-429. doi: 10.1534/genetics.119.302391. Epub 2019 Aug 7. Genetics. 2019. PMID: 31391264 Free PMC article. - Telomere length homeostasis.
Hug N, Lingner J. Hug N, et al. Chromosoma. 2006 Dec;115(6):413-25. doi: 10.1007/s00412-006-0067-3. Epub 2006 Jun 2. Chromosoma. 2006. PMID: 16741708 Review. - Roles of Telomere Biology in Cell Senescence, Replicative and Chronological Ageing.
Liu J, Wang L, Wang Z, Liu JP. Liu J, et al. Cells. 2019 Jan 15;8(1):54. doi: 10.3390/cells8010054. Cells. 2019. PMID: 30650660 Free PMC article. Review.
Cited by
- TERRA and Telomere Maintenance in the Yeast Saccharomyces cerevisiae.
Zeinoun B, Teixeira MT, Barascu A. Zeinoun B, et al. Genes (Basel). 2023 Feb 28;14(3):618. doi: 10.3390/genes14030618. Genes (Basel). 2023. PMID: 36980890 Free PMC article. Review. - Characterization and organization of telomeric-linked helicase (tlh) gene families in Fusarium oxysporum.
Salimi S, Abdi MF, Rahnama M. Salimi S, et al. Curr Genet. 2024 Nov 12;70(1):19. doi: 10.1007/s00294-024-01303-8. Curr Genet. 2024. PMID: 39528830 - Subtelomeric Transcription and its Regulation.
Kwapisz M, Morillon A. Kwapisz M, et al. J Mol Biol. 2020 Jul 10;432(15):4199-4219. doi: 10.1016/j.jmb.2020.01.026. Epub 2020 Feb 6. J Mol Biol. 2020. PMID: 32035903 Free PMC article. Review. - Tea Polyphenols as Prospective Natural Attenuators of Brain Aging.
Hong M, Yu J, Wang X, Liu Y, Zhan S, Wu Z, Zhang X. Hong M, et al. Nutrients. 2022 Jul 22;14(15):3012. doi: 10.3390/nu14153012. Nutrients. 2022. PMID: 35893865 Free PMC article. Review. - Architecture and evolution of subtelomeres in the unicellular green alga Chlamydomonas reinhardtii.
Chaux-Jukic F, O'Donnell S, Craig RJ, Eberhard S, Vallon O, Xu Z. Chaux-Jukic F, et al. Nucleic Acids Res. 2021 Jul 21;49(13):7571-7587. doi: 10.1093/nar/gkab534. Nucleic Acids Res. 2021. PMID: 34165564 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases