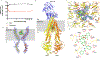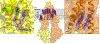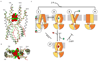Structural insight into substrate and inhibitor discrimination by human P-glycoprotein - PubMed (original) (raw)
Structural insight into substrate and inhibitor discrimination by human P-glycoprotein
Amer Alam et al. Science. 2019.
Abstract
ABCB1, also known as P-glycoprotein, actively extrudes xenobiotic compounds across the plasma membrane of diverse cells, which contributes to cellular drug resistance and interferes with therapeutic drug delivery. We determined the 3.5-angstrom cryo-electron microscopy structure of substrate-bound human ABCB1 reconstituted in lipidic nanodiscs, revealing a single molecule of the chemotherapeutic compound paclitaxel (Taxol) bound in a central, occluded pocket. A second structure of inhibited, human-mouse chimeric ABCB1 revealed two molecules of zosuquidar occupying the same drug-binding pocket. Minor structural differences between substrate- and inhibitor-bound ABCB1 sites are amplified toward the nucleotide-binding domains (NBDs), revealing how the plasticity of the drug-binding site controls the dynamics of the adenosine triphosphate-hydrolyzing NBDs. Ordered cholesterol and phospholipid molecules suggest how the membrane modulates the conformational changes associated with drug binding and transport.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
Fig. 1
In vitro function and structure of nanodisc-reconstituted ABCB1. A Taxol-and zosuquidar-modulated ATPase activity (n=3, error bars indicate SD). B Ribbon diagram of human ABCB1 bound to taxol (green spheres). The N-and C-terminal halves of ABCB1 are colored yellow and orange, respectively with the UIC2 Fab shown in in blue. C Close up of binding site showing side chains of residues within 5Å of bound taxol (green sticks), viewed parallel to the membrane plane. EM density is shown as a blue mesh, contoured at 6 σ. D Ribbon representation of TM4 (yellow) and TM10 (orange) adopting kinked conformation, with taxol located in the center of the occluded cavity. EM density after nanodisc subtraction is contoured at 8 σ. E interactions between taxol and ABCB1 side chains. Non-bonded interactions are represented by spoked arcs and hydrogen bonds are indicated by dashed black lines.
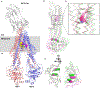
Figure 2.
Comparison of zosuquidar-and taxol-bound ABCB1. A Cartoon of zosuquidar-bound ABCB1HM-EQ structure with-zosuquidar molecules shown as yellow and magenta spheres. The N-and C-terminal halves of ABCB1 are colored pink and blue. B-D Superposition of taxol-bound human ABCB1 (green ribbon) and zosuquidar-bound ABCB1HM-EQ (magenta ribbon) with intracellular helices interacting with NBDs shown as cylinders. Zosuquidar molecules are shown in sphere representation.
Figure 3
Phospholipids and cholesterol bound to ABCB1. Center panel: Surface representation of taxol-bound human ABCB1 showing bound lipid (PE, magenta spheres) and cholesterol molecules (purple spheres). Zoom in panels show details of binding sites of phospholipid (left panel, magenta sticks) and cholesterol (right panel, purple sticks) at the level of the inner leaflet, near the kinking TM helices TM4 and TM10. EM density (blue mesh) is contoured at 6σ.
Figure 4
Proposed mechanism of P-glycoprotein / ABCB1 substrate transport and small-molecule inhibition. A Side view of select TM helices of ABCB1 after superposition of the drug-bound, occluded conformation (green and magenta ribbons for taxol-and zosuquidar-bound structures) with the previously reported, ATP-bound post-translocation state (dark grey ribbon). Taxol and zosuquidar molecules are shown as green and red spheres, respectively. B Same as A but viewed from the cytoplasm. C Schematic of proposed ABCB1 transport cycle in the presence of substrate (taxol, green star) and inhibitor (zosuquidar, red L-shape). ATP and ADP are indicated by T and D, respectively. Dashed lines in models represent ATP binding elements required for NBD dimerization and ATP hydrolysis. Major conformational states are represented by circled numbers. State 1: Apo state (pdb IDs 4M1M & 4QNH, among others); States 2 and 2’: Drug-and inhibitor-bound, this study. State 3: Proposed outward-facing conformation based on Sav1866 structure(30). State 4: Collapsed post-translocation state (pdb ID 6C0V).
Similar articles
- Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Alam A, Küng R, Kowal J, McLeod RA, Tremp N, Broude EV, Roninson IB, Stahlberg H, Locher KP. Alam A, et al. Proc Natl Acad Sci U S A. 2018 Feb 27;115(9):E1973-E1982. doi: 10.1073/pnas.1717044115. Epub 2018 Feb 13. Proc Natl Acad Sci U S A. 2018. PMID: 29440498 Free PMC article. - Mechanism of allosteric modulation of P-glycoprotein by transport substrates and inhibitors.
Dastvan R, Mishra S, Peskova YB, Nakamoto RK, Mchaourab HS. Dastvan R, et al. Science. 2019 May 17;364(6441):689-692. doi: 10.1126/science.aav9406. Science. 2019. PMID: 31097669 Free PMC article. - Drug-protein hydrogen bonds govern the inhibition of the ATP hydrolysis of the multidrug transporter P-glycoprotein.
Chufan EE, Kapoor K, Ambudkar SV. Chufan EE, et al. Biochem Pharmacol. 2016 Feb 1;101:40-53. doi: 10.1016/j.bcp.2015.12.007. Epub 2015 Dec 11. Biochem Pharmacol. 2016. PMID: 26686578 Free PMC article. - ABCB1/MDR1/P-gp employs an ATP-dependent twist-and-squeeze mechanism to export hydrophobic drugs.
Kodan A, Futamata R, Kimura Y, Kioka N, Nakatsu T, Kato H, Ueda K. Kodan A, et al. FEBS Lett. 2021 Mar;595(6):707-716. doi: 10.1002/1873-3468.14018. Epub 2020 Dec 11. FEBS Lett. 2021. PMID: 33275773 Review. - P-glycoprotein-mediated resistance to chemotherapy in cancer cells: using recombinant cytosolic domains to establish structure-function relationships.
Di Pietro A, Dayan G, Conseil G, Steinfels E, Krell T, Trompier D, Baubichon-Cortay H, Jault J. Di Pietro A, et al. Braz J Med Biol Res. 1999 Aug;32(8):925-39. doi: 10.1590/s0100-879x1999000800001. Braz J Med Biol Res. 1999. PMID: 10454753 Review.
Cited by
- A structural framework for unidirectional transport by a bacterial ABC exporter.
Fan C, Kaiser JT, Rees DC. Fan C, et al. Proc Natl Acad Sci U S A. 2020 Aug 11;117(32):19228-19236. doi: 10.1073/pnas.2006526117. Epub 2020 Jul 23. Proc Natl Acad Sci U S A. 2020. PMID: 32703810 Free PMC article. - Theoretical insights on helix repacking as the origin of P-glycoprotein promiscuity.
Bonito CA, Ferreira RJ, Ferreira MU, Gillet JP, Cordeiro MNDS, Dos Santos DJVA. Bonito CA, et al. Sci Rep. 2020 Jun 17;10(1):9823. doi: 10.1038/s41598-020-66587-5. Sci Rep. 2020. PMID: 32555203 Free PMC article. - Mechanistic Insights into Photodynamic Regulation of Adenosine 5'-Triphosphate-Binding Cassette Drug Transporters.
Liang BJ, Lusvarghi S, Ambudkar SV, Huang HC. Liang BJ, et al. ACS Pharmacol Transl Sci. 2021 Sep 15;4(5):1578-1587. doi: 10.1021/acsptsci.1c00138. eCollection 2021 Oct 8. ACS Pharmacol Transl Sci. 2021. PMID: 36118950 Free PMC article. - Molecular Modeling Studies to Probe the Binding Hypothesis of Novel Lead Compounds against Multidrug Resistance Protein ABCB1.
Cheema Y, Linton KJ, Jabeen I. Cheema Y, et al. Biomolecules. 2024 Jan 16;14(1):114. doi: 10.3390/biom14010114. Biomolecules. 2024. PMID: 38254714 Free PMC article. - Phylogenetic analysis of ABCG subfamily proteins in plants: functional clustering and coevolution with ABCGs of pathogens.
Cho CH, Jang S, Choi BY, Hong D, Choi DS, Choi S, Kim H, Han SK, Kim S, Kim MS, Palmgren M, Sohn KH, Yoon HS, Lee Y. Cho CH, et al. Physiol Plant. 2021 Jul;172(3):1422-1438. doi: 10.1111/ppl.13052. Epub 2020 Apr 6. Physiol Plant. 2021. PMID: 31828796 Free PMC article.
References
- Fromm MF, Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol Sci 25, 423–429 (2004). - PubMed
- Borst P, Elferink RO, Mammalian ABC transporters in health and disease. Annu Rev Biochem 71, 537–592 (2002). - PubMed
- Loscher W, Potschka H, Drug resistance in brain diseases and the role of drug efflux transporters. Nat Rev Neurosci 6, 591–602 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases