Challenges and opportunities in cryo-EM single-particle analysis - PubMed (original) (raw)
Review
Challenges and opportunities in cryo-EM single-particle analysis
Dmitry Lyumkis. J Biol Chem. 2019.
Abstract
Cryogenic electron microscopy (cryo-EM) enables structure determination of macromolecular objects and their assemblies. Although the techniques have been developing for nearly four decades, they have gained widespread attention in recent years due to technical advances on numerous fronts, enabling traditional microscopists to break into the world of molecular structural biology. Many samples can now be routinely analyzed at near-atomic resolution using standard imaging and image analysis techniques. However, numerous challenges to conventional workflows remain, and continued technical advances open entirely novel opportunities for discovery and exploration. Here, I will review some of the main methods surrounding cryo-EM with an emphasis specifically on single-particle analysis, and I will highlight challenges, open questions, and opportunities for methodology development.
Keywords: atomic resolution; cryo-electron microscopy; protein structure; protein–protein interaction; single-particle analysis; structural biology.
© 2019 Lyumkis.
Conflict of interest statement
The author declares that he has no conflict of interest with the contents of this article
Figures
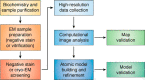
Figure 1.
General workflow for single-particle analysis. The main steps in the SPA workflow are depicted and will be referred to throughout the text. Although the workflow is depicted as approximately linear, often times the process is iterative, and it may be necessary to go back and optimize individual steps prior to proceeding forward.
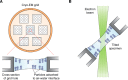
Figure 2.
Preferred particle orientation and specimen tilting. A, top view of a cryo-EM grid (schematic) and selected cross-section within a particular hole. Particles within each hole are typically adsorbed to one of two air–water interfaces, causing them to adopt a preferred orientation on grids (87). Preferred particle orientation leads to anisotropic resolution in the reconstructed map. B, to overcome the preferred orientation and anisotropic resolution problem, the grid can be tilted inside the electron microscope (electron beam is in green). This results in more even coverage of Fourier space voxels and an improvement in the reconstructed volume.
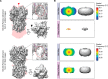
Figure 3.
Experimental results showing the utility of a tilted data collection strategy. Comparison of HA trimer reconstructions refined and reconstructed independently from 0°-untilted images (top) or from 40°-tilted images (bottom). A, left to right, side view of the experimental reconstruction (in gray, the direction of preferred orientation is indicated by the red arrowhead) superimposed onto a projection of the envelope of the HA trimer crystal structure (in pink), displayed alongside a top view and a close-up of a particular region. The black arrowhead indicates streaking in the unsharpened map, which results from misalignment of orientations and/or overfitting. B, experimental half-map and map-to-model 3D FSC isosurfaces. The map-to-model 3D FSCs, which measure correlation to the true structure, are much worse than the half-map 3D FSCs, which measure the internal correlation of two randomly selected half-subsets of the data during refinement. This indicates that some amount of overfitting is present during refinement, since the resulting maps are worse than their apparent reported resolution. Figure adapted from Ref. .
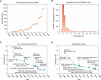
Figure 4.
Advances in cryo-EM map depositions and resolutions by year. A, new depositions into the Electron Microscopy Data Bank by year. B, resolutions of deposited maps in 2018. C, highest resolutions for non-icosahedral specimens by year. D, highest resolutions for icosahedral specimens by year.
Figure 5.
High-resolution cryo-EM maps. A, β-gal, resolved to 1.90 Å (125). B, adeno-associated virus type 2, resolved to 1.86 Å (the reconstruction indicated the presence of hydrogen atoms) (124). C, human apoferritin, resolved to 1.65 Å (126). D, mouse apoferritin, resolved to 1.62 Å (R. Danev, H. Yanagisawa, and M. Kikkawa, manuscript in preparation), represent the state-of-the-art in cryo-EM reconstructions. The codes (EMD-####) refer to the deposition numbers within the
e
lectron
m
icroscopy
d
atabank (EMD).
Figure 6.
Validation procedures for cryo-EM maps and models. Steps for helping to ensure correctness of the derived cryo-EM map (A) and associated atomic model (B). Specific recommendations can be found in Refs. , .
Similar articles
- Visualization of biological macromolecules at near-atomic resolution: cryo-electron microscopy comes of age.
Mitra AK. Mitra AK. Acta Crystallogr F Struct Biol Commun. 2019 Jan 1;75(Pt 1):3-11. doi: 10.1107/S2053230X18015133. Epub 2019 Jan 1. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 30605120 Free PMC article. Review. - Single-particle cryo-electron microscopy of macromolecular complexes.
Skiniotis G, Southworth DR. Skiniotis G, et al. Microscopy (Oxf). 2016 Feb;65(1):9-22. doi: 10.1093/jmicro/dfv366. Epub 2015 Nov 25. Microscopy (Oxf). 2016. PMID: 26611544 Free PMC article. Review. - A primer to single-particle cryo-electron microscopy.
Cheng Y, Grigorieff N, Penczek PA, Walz T. Cheng Y, et al. Cell. 2015 Apr 23;161(3):438-449. doi: 10.1016/j.cell.2015.03.050. Cell. 2015. PMID: 25910204 Free PMC article. Review. - Sample preparation of biological macromolecular assemblies for the determination of high-resolution structures by cryo-electron microscopy.
Stark H, Chari A. Stark H, et al. Microscopy (Oxf). 2016 Feb;65(1):23-34. doi: 10.1093/jmicro/dfv367. Epub 2015 Dec 15. Microscopy (Oxf). 2016. PMID: 26671943 Review. - Refinement of Atomic Structures Against cryo-EM Maps.
Murshudov GN. Murshudov GN. Methods Enzymol. 2016;579:277-305. doi: 10.1016/bs.mie.2016.05.033. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27572731 Review.
Cited by
- Uncovering structural ensembles from single-particle cryo-EM data using cryoDRGN.
Kinman LF, Powell BM, Zhong ED, Berger B, Davis JH. Kinman LF, et al. Nat Protoc. 2023 Feb;18(2):319-339. doi: 10.1038/s41596-022-00763-x. Epub 2022 Nov 14. Nat Protoc. 2023. PMID: 36376590 Free PMC article. Review. - Biasing AlphaFold2 to predict GPCRs and kinases with user-defined functional or structural properties.
Sala D, Hildebrand PW, Meiler J. Sala D, et al. Front Mol Biosci. 2023 Feb 16;10:1121962. doi: 10.3389/fmolb.2023.1121962. eCollection 2023. Front Mol Biosci. 2023. PMID: 36876042 Free PMC article. - Interpretation of medium resolution cryoEM maps of multi-protein complexes.
Casañal A, Shakeel S, Passmore LA. Casañal A, et al. Curr Opin Struct Biol. 2019 Oct;58:166-174. doi: 10.1016/j.sbi.2019.06.009. Epub 2019 Jul 27. Curr Opin Struct Biol. 2019. PMID: 31362190 Free PMC article. Review. - A hybrid approach to study large conformational transitions of biomolecules from single particle XFEL diffraction data.
Asi H, Dasgupta B, Nagai T, Miyashita O, Tama F. Asi H, et al. Front Mol Biosci. 2022 Aug 29;9:913860. doi: 10.3389/fmolb.2022.913860. eCollection 2022. Front Mol Biosci. 2022. PMID: 36660427 Free PMC article. - CryoEM analysis of small plant biocatalysts at sub-2 Å resolution.
Dimos N, Helmer CPO, Chánique AM, Wahl MC, Kourist R, Hilal T, Loll B. Dimos N, et al. Acta Crystallogr D Struct Biol. 2022 Jan 1;78(Pt 1):113-123. doi: 10.1107/S205979832101216X. Epub 2022 Jan 1. Acta Crystallogr D Struct Biol. 2022. PMID: 34981767 Free PMC article.
References
- Campbell M. G., Cheng A., Brilot A. F., Moeller A., Lyumkis D., Veesler D., Pan J., Harrison S. C., Potter C. S., Carragher B., and Grigorieff N. (2012) Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20, 1823–1828 10.1016/j.str.2012.08.026 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM132465/GM/NIGMS NIH HHS/United States
- U54 GM103368/GM/NIGMS NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- R01 AI136680/AI/NIAID NIH HHS/United States
- DP5 OD021396/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources