The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome - PubMed (original) (raw)
Review
The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome
Hemant Gujar et al. Genes (Basel). 2019.
Abstract
A DNA sequence is the hard copy of the human genome and it is a driving force in determining the physiological processes in an organism. Concurrently, the chemical modification of the genome and its related histone proteins is dynamically involved in regulating physiological processes and diseases, which overall constitutes the epigenome network. Among the various forms of epigenetic modifications, DNA methylation at the C-5 position of cytosine in the cytosine⁻guanine (CpG) dinucleotide is one of the most well studied epigenetic modifications. DNA methyltransferases (DNMTs) are a family of enzymes involved in generating and maintaining CpG methylation across the genome. In mammalian systems, DNA methylation is performed by DNMT1 and DNMT3s (DNMT3A and 3B). DNMT1 is predominantly involved in the maintenance of DNA methylation during cell division, while DNMT3s are involved in establishing de novo cytosine methylation and maintenance in both embryonic and somatic cells. In general, all DNMTs require accessory proteins, such as ubiquitin-like containing plant homeodomain (PHD) and really interesting new gene (RING) finger domain 1 (UHRF1) or DNMT3-like (DNMT3L), for their biological function. This review mainly focuses on the role of DNMT3B and its isoforms in de novo methylation and maintenance of DNA methylation, especially with respect to their role as an accessory protein.
Keywords: DNA methylation; DNA methyltransferase; DNMT; DNMT3B; epigenetics.
Conflict of interest statement
Daniel J. Weisenberger is a consultant for the Zymo Research Corporation.
Figures
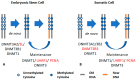
Figure 1
General concept of DNA methylation: (A) DNA methylation in embryonic stem cells (ESCs): ESCs express DNA methyltransferases 3A2 and 3L (DNMT3A2 and DNMT3L), which interact with each other through the carboxy domain. DNMT3L interacts with the unmethylated form of histone H3K4. The catalytically active isoform DNMT3B1 is also highly expressed in embryonic cells. UHRF1 and DNMT1 also maintain DNA methylation in ESCs. (B) DNA methylation in somatic cells: somatic cells mainly express and maintain methylation using DNMT1 and its accessory protein UHRF1. Somatic cells express the catalytically active DNMT3A1 and DNMT3B2 (at low expression levels) that interact with DNMT3B isoforms to catalyze methyl transfer. This interaction is required in the maintenance process. Somatic cells also express the catalytically inactive isoform, DNMT3B3. The black font indicates active DNMT, while the red font indicates an accessary protein.
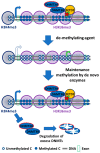
Figure 2
Mechanism of de novo DNA methyltransferases (DNMTs): DNMT3s localizes on methylated cytosine–guanine (CpG)-rich locus to maintain a uniform methylation pattern. DNMT3B targets gene body to methylate cytosine locus. DNMT3B acts as maintenance enzyme, complimenting the low fidelity of DNMT1 and as a de novo enzyme to mark new methylation patterns during differentiation and remethylation after treatment with demethylating agents. DNMT3B also interact with H3K36me3 to localize at these active expression marks. DNA methylation in the genic region is essential for efficient transcription, stability of splicing factors, stability of elongation factor, and to inhibit generation of spurious transcripts. DNMT3s localization at the promoter region is prevented by unmethylated cytosines and by H3K4me3. The free DNMT3s are unstable and are degraded.
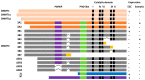
Figure 3
Schematic diagram showing DNA methyltransferase (DNMT) isoforms: DNMT3A (A1 and A2), DNMT3B (3B1, 3B2, 3B3, 3B4, 3B5, 3B6, 3B7, Δ3B1, Δ3B2, Δ3B3, and Δ3B4), and DNMT3L. DNMT3 consist of a PWWP domain (purple), a PHD-like domain (green), and a catalytic domain (black). The deletions are shown as black lines; frame-shift mutations are in yellow. The figure was modified and adapted from Duymich et al., 2016 [113]; Ostler et al., 2007 [110]; Gopalakrishnan et al., 2009 [112]; and Choi et al., 2011 [131]. ESC: Embryonic stem cells.
Similar articles
- DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells.
Duymich CE, Charlet J, Yang X, Jones PA, Liang G. Duymich CE, et al. Nat Commun. 2016 Apr 28;7:11453. doi: 10.1038/ncomms11453. Nat Commun. 2016. PMID: 27121154 Free PMC article. - Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma.
Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Hervouet E, et al. Clin Epigenetics. 2018 Feb 9;10:17. doi: 10.1186/s13148-018-0450-y. eCollection 2018. Clin Epigenetics. 2018. PMID: 29449903 Free PMC article. Review. - The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation.
Zeng Y, Ren R, Kaur G, Hardikar S, Ying Z, Babcock L, Gupta E, Zhang X, Chen T, Cheng X. Zeng Y, et al. Genes Dev. 2020 Nov 1;34(21-22):1546-1558. doi: 10.1101/gad.341925.120. Epub 2020 Oct 1. Genes Dev. 2020. PMID: 33004415 Free PMC article. - DNA Methyltransferases in Mammalian Oocytes.
Uysal F, Ozturk S. Uysal F, et al. Results Probl Cell Differ. 2017;63:211-222. doi: 10.1007/978-3-319-60855-6_10. Results Probl Cell Differ. 2017. PMID: 28779320 Review. - Identification of preferential target sites for human DNA methyltransferases.
Choi SH, Heo K, Byun HM, An W, Lu W, Yang AS. Choi SH, et al. Nucleic Acids Res. 2011 Jan;39(1):104-18. doi: 10.1093/nar/gkq774. Epub 2010 Sep 13. Nucleic Acids Res. 2011. PMID: 20841325 Free PMC article.
Cited by
- What impact does oocyte vitrification have on epigenetics and gene expression?
Barberet J, Barry F, Choux C, Guilleman M, Karoui S, Simonot R, Bruno C, Fauque P. Barberet J, et al. Clin Epigenetics. 2020 Aug 10;12(1):121. doi: 10.1186/s13148-020-00911-8. Clin Epigenetics. 2020. PMID: 32778156 Free PMC article. Review. - Interactions of circRNAs with methylation: An important aspect of circRNA biogenesis and function (Review).
Zhang C, Cui H, Huang C, Kong F, Yang Q, Miao P, Cao Z, Zhang W, Chang D. Zhang C, et al. Mol Med Rep. 2022 May;25(5):169. doi: 10.3892/mmr.2022.12685. Epub 2022 Mar 18. Mol Med Rep. 2022. PMID: 35302170 Free PMC article. - Editorial-Role of DNA Methyltransferases in the Epigenome.
Jeltsch A, Gowher H. Jeltsch A, et al. Genes (Basel). 2019 Jul 30;10(8):574. doi: 10.3390/genes10080574. Genes (Basel). 2019. PMID: 31366147 Free PMC article. - The Common Partner of Several Methyltransferases TRMT112 Regulates the Expression of N6AMT1 Isoforms in Mammalian Cells.
Leetsi L, Õunap K, Abroi A, Kurg R. Leetsi L, et al. Biomolecules. 2019 Aug 28;9(9):422. doi: 10.3390/biom9090422. Biomolecules. 2019. PMID: 31466382 Free PMC article. - Expression of DNMTs and H3K9ac in Ameloblastoma and Ameloblastic Carcinoma.
do Amaral-Silva GK, Morais TML, Wagner VP, Martins MD, Fregnani ER, Soares FA, Rocha AC, Pontes HR, Santos-Silva AR, Vargas PA. do Amaral-Silva GK, et al. Front Oral Health. 2021 Oct 26;2:751162. doi: 10.3389/froh.2021.751162. eCollection 2021. Front Oral Health. 2021. PMID: 35048062 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous