Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism - PubMed (original) (raw)
Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism
Disa Bäckström et al. mBio. 2019.
Abstract
The nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes (proposed order, "Megavirales") include the families Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycodnaviridae, Marseilleviridae, and Mimiviridae, as well as still unclassified pithoviruses, pandoraviruses, molliviruses, and faustoviruses. Several of these virus groups include giant viruses, with genome and particle sizes exceeding those of many bacterial and archaeal cells. We explored the diversity of the NCLDV in deep sea sediments from the Loki's Castle hydrothermal vent area. Using metagenomics, we reconstructed 23 high-quality genomic bins of novel NCLDV, 15 of which are related to pithoviruses, 5 to marseilleviruses, 1 to iridoviruses, and 2 to klosneuviruses. Some of the identified pithovirus-like and marseillevirus-like genomes belong to deep branches in the phylogenetic tree of core NCLDV genes, substantially expanding the diversity and phylogenetic depth of the respective groups. The discovered viruses, including putative giant members of the family Marseilleviridae, have a broad range of apparent genome sizes, in agreement with the multiple, independent origins of gigantism in different branches of the NCLDV. Phylogenomic analysis reaffirms the monophyly of the pithovirus-iridovirus-marseillevirus branch of the NCLDV. Similarly to other giant viruses, the pithovirus-like viruses from Loki's Castle encode translation systems components. Phylogenetic analysis of these genes indicates a greater bacterial contribution than had been detected previously. Genome comparison suggests extensive gene exchange between members of the pithovirus-like viruses and Mimiviridae Further exploration of the genomic diversity of Megavirales in additional sediment samples is expected to yield new insights into the evolution of giant viruses and the composition of the ocean megavirome.IMPORTANCE Genomics and evolution of giant viruses are two of the most vigorously developing areas of virus research. Lately, metagenomics has become the main source of new virus genomes. Here we describe a metagenomic analysis of the genomes of large and giant viruses from deep sea sediments. The assembled new virus genomes substantially expand the known diversity of the nucleocytoplasmic large DNA viruses of eukaryotes. The results support the concept of independent evolution of giant viruses from smaller ancestors in different virus branches.
Keywords: deep sea sediments; giant viruses; metagenomics; nucleocytoplasmic large DNA viruses; virus evolution.
Copyright © 2019 Bäckström et al.
Figures
FIG 1
Diversity of the NCLDV DNAP sequences in the Loki’s Castle sediment metagenomes (orange) and in the Tara Oceans (turquoise) and EarthVirome (purple) databases. Reference sequences are shown in black. The binned NCLDV genomes are marked with a star. Branches with bootstrap values above 95 are marked with a black circle. The maximum likelihood phylogeny was constructed as described in Materials and Methods.
FIG 2
Flowchart of the metagenomic binning procedures. Two main binning approaches were used: differential coverage (DC) binning and coassembly (CA) binning. For DC binning, reads from four different samples were assembled into four metagenomes. The metagenomes were screened for NCLDV DNAP, and contigs were binned with CONCOCT and ESOM. The raw CONCOCT and ESOM bins were combined and refined using mmgenome. The refined bins were put through taxonomic filtering, keeping only the contigs encoding at least one NCLDV gene, and were finally reassembled. For CA binning, a database containing the refined DC bins and NCLDV reference genomes was used to create profiles to extract reads from the metagenomes. The reads were combined and coassembled. This step was followed by CONCOCT binning, mmgenome bin refinement, and taxonomic filtering. Finally, the DC bins and CA bins were annotated and the best bins were chosen by comparing sequence statistics, completeness and redundancy of marker genes, and marker gene phylogenies (see Text S1 for details).
FIG 3
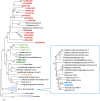
Phylogenetic tree of three concatenated, universally conserved NCLDV proteins: DNA polymerase, major capsid protein, and A18-like helicase. Support values were obtained using 100 bootstrap replications; branches with less than 50% support were collapsed. Scale bars represent the number of amino acid substitutions per site. The inset shows the Mimiviridae branch. Triangles show collapsed branches. The LCV sequences are color-coded as follows: red, pithovirus-like virus; green, marseillevirus-like virus (a deep branch is shown in dark green); orange, iridovirus-like virus; blue, mimivirus (klosneuvirus)-like virus.
FIG 4
Phylogenies of selected translation system components encoded by Loki’s Castle viruses. (A) Translation initiation factor eIF2b. (B) Aspartyl/asparaginyl-tRNA synthetase (AsnS). (C) Tyrosyl-tRNA synthetase (TyrS). (D) Methionyl-tRNA synthetase (MetS). All branches are color-coded according to taxonomic affinity (see Text S2 for the full trees). The numbers at the internal branches indicate (percent) local likelihood-based support.
FIG 5
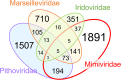
Shared and unique genes in four NCLDV families that include Loki’s Castle viruses. The numbers correspond to NCLDV clusters that contain at least one protein from Mimiviridae, Marseilleviridae, Pithoviridae, and Iridoviridae but are absent from other NCLDV families.
FIG 6
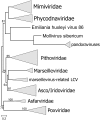
Gene presence-absence tree of the NCLDV that include the Loki’s Castle viruses. The neighbor-joining dendrogram was reconstructed from the matrix of pairwise distances calculated from binary phyletic patterns of the NCLDV clusters. The numbers at internal branches indicate (percent) bootstrap support; data below 50% are not shown.
FIG 7
Phylogenies of selected repair and nucleotide metabolism genes of the pithovirus-iridovirus-marseillevirus group that includes Loki’s Castle viruses. (A) SbcCD nuclease, ATPase subunit SbcC. (B) SbcCD nuclease, nuclease subunit SbcD. (C) Exonuclease V. (D) DNMP kinase. The numbers at the internal branches indicate (percent) local likelihood-based support. GenBank protein identifiers (IDs), wherever available, are shown after each “@” symbol. Taxon abbreviations are as follows: A, Archaea; B, Bacteria; E, Eukaryotes; N, NCLDV; DP, DPANN group; TA, Thaumarchaeota; Ea, Euryarchaeota; FC, Bacteroidetes; Fu, Fusobacteria; Pr, Proteobacteria; Te, Firmicutes; un, unclassified Bacteria; Op, Opisthokonta; Pi, “Pithoviridae”; Ac, Ascoviridae; As, Asfarviridae; Ma, Marseilleviridae; Mi, Mimiviridae; Pa, Pandoraviridae; Ph, Phycodnaviridae; V ds, double-strand DNA viruses.
FIG 8
Loki’s Castle virophages. (A) Phylogenetic tree of virophage major capsid proteins. Reference virophages from GenBank are marked with black font (the three prototype virophages are shown in bold); environmental virophages are shown in blue (129) and green (wgs portion of GenBank). (B) Genome maps of Loki’s Castle virophages compared with Sputnik virophage. Green and blue triangles mark direct and inverted repeats. Pentagons with a thick outline represent conserved virophage genes.
Similar articles
- Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.
Koonin EV, Yutin N. Koonin EV, et al. Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review. - Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life.
Yutin N, Wolf YI, Koonin EV. Yutin N, et al. Virology. 2014 Oct;466-467:38-52. doi: 10.1016/j.virol.2014.06.032. Epub 2014 Jul 17. Virology. 2014. PMID: 25042053 Free PMC article. - Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.
Christo-Foroux E, Alempic JM, Lartigue A, Santini S, Labadie K, Legendre M, Abergel C, Claverie JM. Christo-Foroux E, et al. J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article. - Evidence of the megavirome in humans.
Colson P, Fancello L, Gimenez G, Armougom F, Desnues C, Fournous G, Yoosuf N, Million M, La Scola B, Raoult D. Colson P, et al. J Clin Virol. 2013 Jul;57(3):191-200. doi: 10.1016/j.jcv.2013.03.018. Epub 2013 May 8. J Clin Virol. 2013. PMID: 23664726 - Updating strategies for isolating and discovering giant viruses.
Khalil JYB, Andreani J, La Scola B. Khalil JYB, et al. Curr Opin Microbiol. 2016 Jun;31:80-87. doi: 10.1016/j.mib.2016.03.004. Epub 2016 Apr 1. Curr Opin Microbiol. 2016. PMID: 27039269 Review.
Cited by
- A Virome and Proteomic Analysis of Placental Microbiota in Pregnancies with and without Fetal Growth Restriction.
Stupak A, Kwiatek M, Gęca T, Kwaśniewska A, Mlak R, Nawrot R, Goździcka-Józefiak A, Kwaśniewski W. Stupak A, et al. Cells. 2024 Oct 23;13(21):1753. doi: 10.3390/cells13211753. Cells. 2024. PMID: 39513860 Free PMC article. - Global marine microbial diversity and its potential in bioprospecting.
Chen J, Jia Y, Sun Y, Liu K, Zhou C, Liu C, Li D, Liu G, Zhang C, Yang T, Huang L, Zhuang Y, Wang D, Xu D, Zhong Q, Guo Y, Li A, Seim I, Jiang L, Wang L, Lee SMY, Liu Y, Wang D, Zhang G, Liu S, Wei X, Yue Z, Zheng S, Shen X, Wang S, Qi C, Chen J, Ye C, Zhao F, Wang J, Fan J, Li B, Sun J, Jia X, Xia Z, Zhang H, Liu J, Zheng Y, Liu X, Wang J, Yang H, Kristiansen K, Xu X, Mock T, Li S, Zhang W, Fan G. Chen J, et al. Nature. 2024 Sep;633(8029):371-379. doi: 10.1038/s41586-024-07891-2. Epub 2024 Sep 4. Nature. 2024. PMID: 39232160 Free PMC article. - Virophages, Satellite Viruses, Virophage Replication and Its Effects and Virophage Defence Mechanisms for Giant Virus Hosts and Giant Virus Defence Systems against Virophages.
Tokarz-Deptuła B, Chrzanowska S, Baraniecki Ł, Gurgacz N, Stosik M, Sobolewski J, Deptuła W. Tokarz-Deptuła B, et al. Int J Mol Sci. 2024 May 28;25(11):5878. doi: 10.3390/ijms25115878. Int J Mol Sci. 2024. PMID: 38892066 Free PMC article. Review. - Giant viral signatures on the Greenland ice sheet.
Perini L, Sipes K, Zervas A, Bellas C, Lutz S, Moniruzzaman M, Mourot R, Benning LG, Tranter M, Anesio AM. Perini L, et al. Microbiome. 2024 May 17;12(1):91. doi: 10.1186/s40168-024-01796-y. Microbiome. 2024. PMID: 38760842 Free PMC article. - The genomic and phylogenetic analysis of Marseillevirus cajuinensis raises questions about the evolution of Marseilleviridae lineages and their taxonomical organization.
de Azevedo BL, Queiroz VF, de Aquino ILM, Machado TB, de Assis FL, Reis E, Araújo Júnior JP, Ullmann LS, Colson P, Greub G, Aylward F, Rodrigues RAL, Abrahão JS. de Azevedo BL, et al. J Virol. 2024 Jun 13;98(6):e0051324. doi: 10.1128/jvi.00513-24. Epub 2024 May 16. J Virol. 2024. PMID: 38752754 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources