Control of proliferation in the haploid meristem by CLE peptide signaling in Marchantia polymorpha - PubMed (original) (raw)
Control of proliferation in the haploid meristem by CLE peptide signaling in Marchantia polymorpha
Yuki Hirakawa et al. PLoS Genet. 2019.
Abstract
The homeostasis of meristems in flowering plants is maintained by cell-to-cell communication via CLE (CLAVATA3/EMBRYO SURROUNDING REGION-related) peptide hormones. In contrast, cell signals that regulate meristem activity remains elusive in bryophytes that maintain apical meristems in the gametophyte (haploid) body and undergo a gametophyte-dominant life cycle. We here show that MpCLE1 confines the proliferative activity of gametophytic meristem and affects the overall size of gametangiophores (reproductive organs) in Marchantia polymorpha, which is in sharp contrast with the meristem-promoting function of its ortholog TDIF/CLE41/CLE44 in Arabidopsis vascular meristems. Expression analysis suggests that MpCLE1 and its receptor gene MpTDR are expressed in distinct patterns across the apical meristem. These data suggest that local CLE peptide signaling may have had a role in regulating cell proliferation in the shoot meristem in the ancestral land plant and acts in both sporophytic and gametophytic meristems of extant plants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
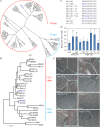
Fig 1. MpCLE1 peptide variants mimic Arabidopsis CLE41/TDIF.
(A) A phylogenetic tree of CLE peptides based on their 12 amino-acid CLE peptide sequences. Two subgroups R-type and H-type are indicated. (B) A phylogenetic tree of CLE peptide receptors based on their kinase domain sequences. TDR and CLV1 subclades are indicated. ER(ERECTA) is set as the outgroup. The posterior probabilities of trees are shown at the nodes in (A) and (B). At, Arabidopsis thaliana; Sm, Selaginella moellendorffii; Nsp, Nothoceros sp.; Pp, Physcomitrella patens; Sl, Sphagnum lescurii; Me, Marchantia emarginata; Mp, Marchantia polymorpha; Rn, Ricciocarpos natans; St, Sphaerocarpos texanus. (C) Peptides used in the assays. Blue characters indicate residues different from the MpCLE1 peptide. Residues “O” indicate hydroxyprolines. (D) Effects of 10 μM peptide treatment on stele thickening in the hypocotyls of 10-day-old Arabidopsis. Peptides are indicated below. “Hyp” and “-” indicate the peptides with and without hyrdoxyprolines, respectively. Note that AtCLE41-Hyp is identical to TDIF. Data represent mean values ± s.d. (n = 13–16). Asterisks indicate a significant difference from mock treatment (0 M) in Welch’s _t_-test, p<0.05. (E) Effects of 10 μM peptide treatment in the leaf vein of 10-day-old Arabidopsis. Red and cyan lines indicate vein and xylem strand, respectively. Scale bars = 100um.
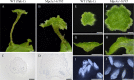
Fig 2. Gain-of-function phenotypes of Mp_CLE1_.
Morphology of 14-day-old M. polymorpha plants grown from gemmae. (A and B) Overall morphology of (A) wild type (Tak-1) and (B) Mp_EF1α_ promoter-driven Mp_CLE1_-overexpression line. (C) Area of thalli (mean ± s.d.). Two independent transgenic lines were used for quantification. Asterisks indicate a significant difference from WT in Weltch’s _t_-test (p < 0.001, n = 18). (D) Schematic diagram of section planes in apical notch. Longitudinal sections of apical notches in (E) wild type (Tak-1) and (F) the Mp_CLE1_-overexpression line. Arrows, red and black arrowheads, and brackets indicate developing gemmae cups, apical cells, developing air chambers closest to the apical cell, and proliferative region of meristem, respectively. Scale bars = 1cm in (A and B), 200 μm in (E, F).
Fig 3. Reduction of meristem activity by Mp_CLE1_.
Consecutive transverse sections of apical notches in 14-day-old plants grown from gemmae. WT (A1-A7) and Mp_CLE1_ overexpression (B1-B7) plants are compared. The relative position (μm) is indicated at the right bottom corner of each panel. Note that “0 μm” (A2 and B2) is set at the section in which two flanking lobes merged in the consecutive transverse sectioning as illustrated in (C). Arrows indicate a gemmae cup. Scale bars = 200 μm.
Fig 4. Loss-of-function phenotypes of Mp_CLE1_ in apical notch.
(A and B) Longitudinal sections of apical notches in 14-day-old M. polymorpha plants grown from gemmae in (A) homologous recombination-based Mp_cle1_ knock-out line and (B) its complementation line. (C and D) Transverse sections of apical notches at “20 μm” position from the junction of two flanking lobes. Note that consecutive sections including these figures are shown in S6 Fig. Red and black arrowheads, and brackets indicate apical cells, developing air chambers closest to the apical cell, and proliferative region of meristem, respectively. Scale bars = 200 μm.
Fig 5. Loss-of-function phenotypes of Mp_CLE1_ in antheridiophore.
Comparison of antheridiophore morphology between WT (A, C, E, G and I) and Mp_cle1_ knock-out line (B, D, F, H and J). (A and B) Overall morphology. (C and D) Stalk cross-sections. (E and F) Antheridial receptacles. (G and H) Hand sections of the antheridial receptacles. (I and J) Antheridia. Scale bars = 2 mm in (A, B and E-H), 1 mm in (I) and (J) and 200 μm in (C) and (D).
Fig 6. Genetic interaction of Mp_CLE1_ and Mp_TDR_. Mp_tdr_ knock-out mutant is insensitive to excess MpCLE1/TDIF.
(A and B) Overall morphology of 21-day-old plants grown from gemmae. (C) Area of thalli in 14-day-old plants (mean ± s.d.). No significant difference was found between Mp_tdr_ and Mp_tdr_ Mp_CLE1ox_ in Weltch’s t_-test (p = 0.19, n = 17–18). (D-G) Overall morphology of 17-day-old plants grown from gemmae with or without 10 μM TDIF. (H-K) Archegonial receptacles. Mp_cle1 and Mp_tdr_ indicate knock-out mutants. Mp_CLE1ox_ indicates Mp_EF1α_ promoter-driven overexpression. Note that Mp_CLE1_ overexpression plants do not make gametangiophores (S3G Fig). Scale bars = 1 cm in (A,B,D-G) and 2 mm in (H-K).
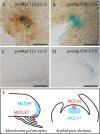
Fig 7. Expression patterns of Mp_CLE1_ and Mp_TDR_ in apical notch.
(A and B) Promoter-GUS reporter assays for Mp_CLE1_ and Mp_TDR_ expression in 5-day-old gemmalings. (C and D) Longitudinal sections of the promoter:GUS lines at the notch in 5-day-old gemmalings. (E) Schematic illustration of promoter activities of CLE (red) and receptor (cyan) genes in the meristem. M. polymorpha (left) genes are contrasted with A. thaliana (right) genes. Scale bars = 200 μm (A and B), 100 μm (C and D).
Similar articles
- Induction of Multichotomous Branching by CLAVATA Peptide in Marchantia polymorpha.
Hirakawa Y, Fujimoto T, Ishida S, Uchida N, Sawa S, Kiyosue T, Ishizaki K, Nishihama R, Kohchi T, Bowman JL. Hirakawa Y, et al. Curr Biol. 2020 Oct 5;30(19):3833-3840.e4. doi: 10.1016/j.cub.2020.07.016. Epub 2020 Aug 20. Curr Biol. 2020. PMID: 32822612 - Control of stem cell behavior by CLE-JINGASA signaling in the shoot apical meristem in Marchantia polymorpha.
Takahashi G, Kiyosue T, Hirakawa Y. Takahashi G, et al. Curr Biol. 2023 Dec 4;33(23):5121-5131.e6. doi: 10.1016/j.cub.2023.10.054. Epub 2023 Nov 16. Curr Biol. 2023. PMID: 37977139 - MpANT regulates meristem development in Marchantia polymorpha.
Liu W, Yang Z, Cai G, Li B, Liu S, Willemsen V, Xu L. Liu W, et al. Cell Rep. 2024 Jul 23;43(7):114466. doi: 10.1016/j.celrep.2024.114466. Epub 2024 Jul 9. Cell Rep. 2024. PMID: 38985681 - CLE peptides: critical regulators for stem cell maintenance in plants.
Song XF, Hou XL, Liu CM. Song XF, et al. Planta. 2021 Nov 29;255(1):5. doi: 10.1007/s00425-021-03791-1. Planta. 2021. PMID: 34841457 Review. - CLE peptide ligands and their roles in establishing meristems.
Fiers M, Ku KL, Liu CM. Fiers M, et al. Curr Opin Plant Biol. 2007 Feb;10(1):39-43. doi: 10.1016/j.pbi.2006.11.003. Epub 2006 Nov 28. Curr Opin Plant Biol. 2007. PMID: 17129751 Review.
Cited by
- Spatial range, temporal span, and promiscuity of CLE-RLK signaling.
Narasimhan M, Simon R. Narasimhan M, et al. Front Plant Sci. 2022 Aug 26;13:906087. doi: 10.3389/fpls.2022.906087. eCollection 2022. Front Plant Sci. 2022. PMID: 36092449 Free PMC article. Review. - Genome-Wide Identification and Characterization of CLAVATA3/EMBRYO SURROUNDING REGION (CLE) Gene Family in Foxtail Millet (Setaria italica L.).
Ren X, Chen J, Chen S, Zhang H, Li L. Ren X, et al. Genes (Basel). 2023 Nov 6;14(11):2046. doi: 10.3390/genes14112046. Genes (Basel). 2023. PMID: 38002989 Free PMC article. - An Evolutionarily Conserved Coreceptor Gene Is Essential for CLAVATA Signaling in Marchantia polymorpha.
Takahashi G, Betsuyaku S, Okuzumi N, Kiyosue T, Hirakawa Y. Takahashi G, et al. Front Plant Sci. 2021 Apr 13;12:657548. doi: 10.3389/fpls.2021.657548. eCollection 2021. Front Plant Sci. 2021. PMID: 33927741 Free PMC article. - The bryophytes Physcomitrium patens and Marchantia polymorpha as model systems for studying evolutionary cell and developmental biology in plants.
Naramoto S, Hata Y, Fujita T, Kyozuka J. Naramoto S, et al. Plant Cell. 2022 Jan 20;34(1):228-246. doi: 10.1093/plcell/koab218. Plant Cell. 2022. PMID: 34459922 Free PMC article. Review. - WOX going on: CLE peptides in plant development.
Willoughby AC, Nimchuk ZL. Willoughby AC, et al. Curr Opin Plant Biol. 2021 Oct;63:102056. doi: 10.1016/j.pbi.2021.102056. Epub 2021 May 30. Curr Opin Plant Biol. 2021. PMID: 34077886 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources