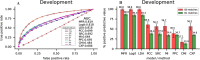Using Machine Learning to Measure Relatedness Between Genes: A Multi-Features Model - PubMed (original) (raw)
Using Machine Learning to Measure Relatedness Between Genes: A Multi-Features Model
Yan Wang et al. Sci Rep. 2019.
Abstract
Measuring conditional relatedness between a pair of genes is a fundamental technique and still a significant challenge in computational biology. Such relatedness can be assessed by gene expression similarities while suffering high false discovery rates. Meanwhile, other types of features, e.g., prior-knowledge based similarities, is only viable for measuring global relatedness. In this paper, we propose a novel machine learning model, named Multi-Features Relatedness (MFR), for accurately measuring conditional relatedness between a pair of genes by incorporating expression similarities with prior-knowledge based similarities in an assessment criterion. MFR is used to predict gene-gene interactions extracted from the COXPRESdb, KEGG, HPRD, and TRRUST databases by the 10-fold cross validation and test verification, and to identify gene-gene interactions collected from the GeneFriends and DIP databases for further verification. The results show that MFR achieves the highest area under curve (AUC) values for identifying gene-gene interactions in the development, test, and DIP datasets. Specifically, it obtains an improvement of 1.1% on average of precision for detecting gene pairs with both high expression similarities and high prior-knowledge based similarities in all datasets, comparing to other linear models and coexpression analysis methods. Regarding cancer gene networks construction and gene function prediction, MFR also obtains the results with more biological significances and higher average prediction accuracy, than other compared models and methods. A website of the MFR model and relevant datasets can be accessed from http://bmbl.sdstate.edu/MFR .
Conflict of interest statement
The authors declare no competing interests.
Figures
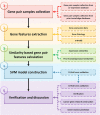
Figure 1
Workflow of MFR model. Five steps are in the workflow, including (i) gene pair samples collection, (ii) gene features extraction, (iii) gene pair features calculation, (iv) SVM model construction and (v) verification and discussion.
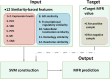
Figure 2
Structure of the MFR model. The model is based on SVM and uses 12 similarity-based gene pair features as input; and the output value, namely MFR, is applied as an assessment criterion for measuring conditional relatedness between genes.
Figure 3
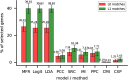
(A) ROCs of nine models or methods for identifying gene-gene interactions by the 10-fold cross-validation. (B) Average PPVs of nine models or methods for detecting B0/B1 matched gene pairs by 10-fold cross-validation.
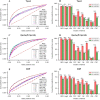
Figure 4
ROCs of nine models or methods for identifying gene-gene interactions in the (A) test, (C) GeneFriends and (E) DIP datasets. Average PPVs of nine models or methods for detecting B0/B1 matched gene pairs in the (B) test, (D) GeneFriends and (F) DIP datasets.
Figure 5
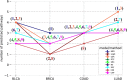
Metabolic pathways are predicted to be directly influenced by increased glutamine and glutamate metabolism in nine BRCA gene networks.
Figure 6
Number of metabolic pathways predicted to be directly influenced by increased glutamine and glutamate metabolism in four cancer types. These pathways were predicted in cancer gene networks, where nodes represent up-regulated metabolic genes and edges represent relatedness between genes, measured by the five linear models and six coexpression analysis methods.
Figure 7
Percentages of L0- and L1-matched selected genes in the nine KEGG metabolic gene networks. In these networks, nodes represent genes involved in KEGG metabolism pathways, and edges represent relatedness between genes, measured by the nine models or methods.
Similar articles
- Measurement of Conditional Relatedness Between Genes Using Fully Convolutional Neural Network.
Wang Y, Zhang S, Yang L, Yang S, Tian Y, Ma Q. Wang Y, et al. Front Genet. 2019 Oct 22;10:1009. doi: 10.3389/fgene.2019.01009. eCollection 2019. Front Genet. 2019. PMID: 31695723 Free PMC article. - Network inference with ensembles of bi-clustering trees.
Pliakos K, Vens C. Pliakos K, et al. BMC Bioinformatics. 2019 Oct 28;20(1):525. doi: 10.1186/s12859-019-3104-y. BMC Bioinformatics. 2019. PMID: 31660848 Free PMC article. - Identifying disease genes using machine learning and gene functional similarities, assessed through Gene Ontology.
Asif M, Martiniano HFMCM, Vicente AM, Couto FM. Asif M, et al. PLoS One. 2018 Dec 10;13(12):e0208626. doi: 10.1371/journal.pone.0208626. eCollection 2018. PLoS One. 2018. PMID: 30532199 Free PMC article. - Systems Biology and Machine Learning in Plant-Pathogen Interactions.
Mishra B, Kumar N, Mukhtar MS. Mishra B, et al. Mol Plant Microbe Interact. 2019 Jan;32(1):45-55. doi: 10.1094/MPMI-08-18-0221-FI. Epub 2018 Nov 12. Mol Plant Microbe Interact. 2019. PMID: 30418085 Review. - LncFinder: an integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property.
Han S, Liang Y, Ma Q, Xu Y, Zhang Y, Du W, Wang C, Li Y. Han S, et al. Brief Bioinform. 2019 Nov 27;20(6):2009-2027. doi: 10.1093/bib/bby065. Brief Bioinform. 2019. PMID: 30084867 Free PMC article. Review.
Cited by
- TopoFun: a machine learning method to improve the functional similarity of gene co-expression modules.
Janbain A, Reynès C, Assaghir Z, Zeineddine H, Sabatier R, Journot L. Janbain A, et al. NAR Genom Bioinform. 2021 Nov 8;3(4):lqab103. doi: 10.1093/nargab/lqab103. eCollection 2021 Dec. NAR Genom Bioinform. 2021. PMID: 34761220 Free PMC article. - DNC4mC-Deep: Identification and Analysis of DNA N4-Methylcytosine Sites Based on Different Encoding Schemes By Using Deep Learning.
Wahab A, Mahmoudi O, Kim J, Chong KT. Wahab A, et al. Cells. 2020 Jul 22;9(8):1756. doi: 10.3390/cells9081756. Cells. 2020. PMID: 32707969 Free PMC article. - Measurement of Conditional Relatedness Between Genes Using Fully Convolutional Neural Network.
Wang Y, Zhang S, Yang L, Yang S, Tian Y, Ma Q. Wang Y, et al. Front Genet. 2019 Oct 22;10:1009. doi: 10.3389/fgene.2019.01009. eCollection 2019. Front Genet. 2019. PMID: 31695723 Free PMC article. - A Bioinformatics Tool for the Prediction of DNA N6-Methyladenine Modifications Based on Feature Fusion and Optimization Protocol.
Cai J, Wang D, Chen R, Niu Y, Ye X, Su R, Xiao G, Wei L. Cai J, et al. Front Bioeng Biotechnol. 2020 Jun 4;8:502. doi: 10.3389/fbioe.2020.00502. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32582654 Free PMC article. - NCResNet: Noncoding Ribonucleic Acid Prediction Based on a Deep Resident Network of Ribonucleic Acid Sequences.
Yang S, Wang Y, Zhang S, Hu X, Ma Q, Tian Y. Yang S, et al. Front Genet. 2020 Feb 28;11:90. doi: 10.3389/fgene.2020.00090. eCollection 2020. Front Genet. 2020. PMID: 32180792 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources