Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes - PubMed (original) (raw)
Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes
Martin Closter Jespersen et al. Front Immunol. 2019.
Abstract
B-cells can neutralize pathogenic molecules by targeting them with extreme specificity using receptors secreted or expressed on their surface (antibodies). This is achieved via molecular interactions between the paratope (i.e., the antibody residues involved in the binding) and the interacting region (epitope) of its target molecule (antigen). Discerning the rules that define this specificity would have profound implications for our understanding of humoral immunogenicity and its applications. The aim of this work is to produce improved, antibody-specific epitope predictions by exploiting features derived from the antigens and their cognate antibodies structures, and combining them using statistical and machine learning algorithms. We have identified several geometric and physicochemical features that are correlated in interacting paratopes and epitopes, used them to develop a Monte Carlo algorithm to generate putative epitopes-paratope pairs, and train a machine-learning model to score them. We show that, by including the structural and physicochemical properties of the paratope, we improve the prediction of the target of a given B-cell receptor. Moreover, we demonstrate a gain in predictive power both in terms of identifying the cognate antigen target for a given antibody and the antibody target for a given antigen, exceeding the results of other available tools.
Keywords: B cell epitope; antibody; antibody specific epitope prediction; antigen; paratope; prediction.
Figures
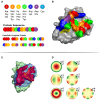
Figure 1
(A) Conjoint Triads amino acid classes and representation of method on a sequence level. (B) Structural representation of Conjoint Triads classes mapped to an epitope patch. (C) The three principal components illustrated on an epitope patch. (D) Illustration of 4th order of Zernike Moments' descriptive shape excluding order 0 and 1.
Figure 2
Correlation matrix of structural and physicochemical features of the true paired paratope and epitope patches.
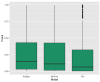
Figure 3
Box plot showing the distribution of the real epitope ranks within each Antibody-Antigen structure for the three prediction models; Antigen, Minimal, and Full.
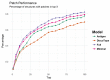
Figure 4
The ability of the four models' (Antigen: green, Minimal: pink, Full: purple, and DiscoTope-2.0: orange) to identify high overlapping patches. X-axis indicating number of top predicted patches included and Y-axis showing the percentage of structures having at least one high overlapping within the selected pool.
Similar articles
- Prediction of Paratope-Epitope Pairs Using Convolutional Neural Networks.
Li D, Pucci F, Rooman M. Li D, et al. Int J Mol Sci. 2024 May 16;25(10):5434. doi: 10.3390/ijms25105434. Int J Mol Sci. 2024. PMID: 38791470 Free PMC article. - Mining for the antibody-antigen interacting associations that predict the B cell epitopes.
Zhao L, Li J. Zhao L, et al. BMC Struct Biol. 2010 May 17;10 Suppl 1(Suppl 1):S6. doi: 10.1186/1472-6807-10-S1-S6. BMC Struct Biol. 2010. PMID: 20487513 Free PMC article. - Correlations between Terasaki's HLA class I epitopes and HLAMatchmaker-defined eplets on HLA-A, -B and -C antigens.
Duquesnoy RJ, Marrari M. Duquesnoy RJ, et al. Tissue Antigens. 2009 Aug;74(2):117-33. doi: 10.1111/j.1399-0039.2009.01271.x. Epub 2009 Jun 2. Tissue Antigens. 2009. PMID: 19497041 - Antigen-Antibody Complexes.
Kapingidza AB, Kowal K, Chruszcz M. Kapingidza AB, et al. Subcell Biochem. 2020;94:465-497. doi: 10.1007/978-3-030-41769-7_19. Subcell Biochem. 2020. PMID: 32189312 Review. - An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction.
Potocnakova L, Bhide M, Pulzova LB. Potocnakova L, et al. J Immunol Res. 2016;2016:6760830. doi: 10.1155/2016/6760830. Epub 2016 Dec 29. J Immunol Res. 2016. PMID: 28127568 Free PMC article. Review.
Cited by
- A novel therapeutic multiepitope vaccine based on oncoprotein E6 and E7 of HPV 16 and 18: An in silico approach.
Pratiwi SE, Ysrafil Y, Mardhia M, Mahyarudin M, Ilmiawan MI, Trianto HF, Liana DF, Amia Y. Pratiwi SE, et al. Bioimpacts. 2024;14(5):27846. doi: 10.34172/bi.2024.27846. Epub 2024 Feb 6. Bioimpacts. 2024. PMID: 39296802 Free PMC article. - Computational prediction of multiple antigen epitopes.
Viswanathan R, Carroll M, Roffe A, Fajardo JE, Fiser A. Viswanathan R, et al. Bioinformatics. 2024 Oct 1;40(10):btae556. doi: 10.1093/bioinformatics/btae556. Bioinformatics. 2024. PMID: 39271143 Free PMC article. - Development of innovative multi-epitope mRNA vaccine against central nervous system tuberculosis using in silico approaches.
Shi H, Zhu Y, Shang K, Tian T, Yin Z, Shi J, He Y, Ding J, Wang Q, Zhang F. Shi H, et al. PLoS One. 2024 Sep 6;19(9):e0307877. doi: 10.1371/journal.pone.0307877. eCollection 2024. PLoS One. 2024. PMID: 39240891 Free PMC article. - Computational Methods to Predict Conformational B-Cell Epitopes.
Carroll M, Rosenbaum E, Viswanathan R. Carroll M, et al. Biomolecules. 2024 Aug 10;14(8):983. doi: 10.3390/biom14080983. Biomolecules. 2024. PMID: 39199371 Free PMC article. Review. - Assessing the predictive ability of computational epitope prediction methods on Fel d 1 and other allergens.
Kwon H, Ko S, Ha K, Lee JK, Choi Y. Kwon H, et al. PLoS One. 2024 Aug 23;19(8):e0306254. doi: 10.1371/journal.pone.0306254. eCollection 2024. PLoS One. 2024. PMID: 39178274 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources