Genome-wide discovery of structured noncoding RNAs in bacteria - PubMed (original) (raw)
Genome-wide discovery of structured noncoding RNAs in bacteria
Shira Stav et al. BMC Microbiol. 2019.
Abstract
Background: Structured noncoding RNAs (ncRNAs) play essential roles in many biological processes such as gene regulation, signaling, RNA processing, and protein synthesis. Among the most common groups of ncRNAs in bacteria are riboswitches. These cis-regulatory, metabolite-binding RNAs are present in many species where they regulate various metabolic and signaling pathways. Collectively, there are likely to be hundreds of novel riboswitch classes that remain hidden in the bacterial genomes that have already been sequenced, and potentially thousands of classes distributed among various other species in the biosphere. The vast majority of these undiscovered classes are proposed to be exceedingly rare, and so current bioinformatics search techniques are reaching their limits for differentiating between true riboswitch candidates and false positives.
Results: Herein, we exploit a computational search pipeline that can efficiently identify intergenic regions most likely to encode structured ncRNAs. Application of this method to five bacterial genomes yielded nearly 70 novel genetic elements including 30 novel candidate ncRNA motifs. Among the riboswitch candidates identified is an RNA motif involved in the regulation of thiamin biosynthesis.
Conclusions: Analysis of other genomes will undoubtedly lead to the discovery of many additional novel structured ncRNAs, and provide insight into the range of riboswitches and other kinds of ncRNAs remaining to be discovered in bacteria and archaea.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
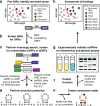
Fig. 1
Overview of the search pipeline. Schematic representation of the GC-IGR analysis workflow. a-h Annotations in bold text represent major steps in the analytical pipeline. For a more detailed version of the pipeline, see Additional file 2: Figure S1
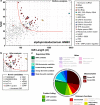
Fig. 2
Plots of the IGRs from the HIMB5 genome sorted based on IGR length and GC content. a IGR plot prior to conducting our detailed analyses. Red line represents the boundary between unknown IGRs chosen for further analysis (upper right) and those that were not chosen (lower left). IGRs selected for further analysis are depicted with dark gray triangles whereas those not chosen are depicted with light gray triangles. b, (Top) A portion of the sector of interest from the HIMB5 IGR plot after analysis, updated to remove false IGRs that overlap known ORFs, to annotate IGRs carrying previously known ncRNAs, and to include all novel motifs identified in this study. No changes in the plot occurred outside of the area depicted. (Bottom) List of novel motif candidates. c, Summary of the classification of all 47 unknown IGRs from HIMB5 chosen for further analysis. Classifications are organized into five main groups (gray arcs) as annotated, wherein “unknown functions” encompasses categories 1 through 4 (unnamed, LRC, MRC and HRC), and the remaining groups are derived from category 5 (named), as described in the main text. The number of novel examples classified in each group are provided in the colored boxes. Classifications depicted as partially transparent lack a representative in the sector of interest in this genome. See Additional file 1: Table S1 for additional details regarding novel motifs
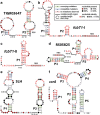
Fig. 3
Sequence and secondary structure models for several candidate riboswitch classes identified in this study. a-d Four candidate riboswitch classes present in the HIMB5 genome. e A candidate riboswitch class present in the genome of T. lienii. f A candidate riboswitch class present in the genome of B. cicadellinicola. These consensus models are created by comparing all unique sequence representatives for each motif that were uncovered by homology searches of RefSeq 76 and certain metagenomic databases. See the text for details regarding each of these motifs and for hypotheses regarding their biological functions. Note that another riboswitch candidate, thiS, identified in the genome of C. novyi is presented in Fig. 4a
Fig. 4
Structure and genetic context of the thiS motif. a Consensus sequence and secondary structure model for the thiS motif. Annotations are as described for Fig. 3. The P0 stem is predicted to exist if the lower portion of P1 fails to form. b (Top) Distribution of gene associations for the ~ 700 thiS motif representatives in bacteria. The chart incorporates the first five genes downstream of the thiS motif, and has a total of 1922 entries. These genes, which are typically in the thiS operon, are counted individually. (Bottom) Protein products of the genes abbreviated here, when known, catalyze the reaction steps for thiamin biosynthesis depicted in c. c The biosynthetic pathway of TPP in Bacillus subtilis. Acronyms starting from the top left are: aminoimidazole ribotide (AIR), hydroxymethyl-pyrimidine (HMP) hydroxymethyl-pyrimidine phosphate (HMP-P), hydroxymethyl-pyrimidine pyrophosphate (HMP-PP), hydroxyethyl-thiazole (HET), hydroxyethyl-thiazole phosphate (HET-P), thiamin monophosphate (TMP), and thiamin pyrophosphate (TPP). TMP (green shaded box) is formed by fusing the two compounds HMP-PP (blue shaded box) and HET-P (gold shaded box). Note that both HET-P and TPP can be synthesized through a salvage pathway starting with HET and thiamin, respectively. HMP-PP and HET-P were proposed as the top ligand candidates for the thiS riboswitch candidate. Metabolic scheme is based on that published previously [56]
Fig. 5
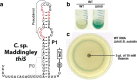
Reporter gene expression is regulated by the thiS riboswitch candidate. a Sequence and predicted secondary structure of the WT thiS RNA associated with the thiS gene of C. maddingley, which was fused to a β-galactosidase reporter gene (lacZ) and a B. subtilis lysC promoter to drive transcription. The lysC promoter was chosen for this purpose because it is known to strongly promote transcription without regulation [73]. Encircled 88 designates the number of additional nucleotides between the end of the terminator element and the lacZ reporter gene sequence. Red nucleotides are > 97% conserved in the thiS consensus model (Fig. 4a). b Reporter gene expression of WT B. subtilis cells and cells lacking the coding region for the ThiS protein (Δ_thiS_) grown in minimal (GMM) liquid media. c Agar diffusion assay of the Δ_thiS B. subtilis_ strain with a WT riboswitch reporter construct. The filter disk was spotted with 10 mM thiamin on a minimal (GMM) agar medium plate with 100 μg mL− 1 X-Gal
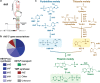
Fig. 6
Representatives of various types of predicted structured nucleic acid motifs discovered among five bacterial genomes. a-e Sequence and predicted secondary structure models for representative ‘named’ motifs identified among the five bacterial genomes examined in this study. Extended blue shading in a and b designate possible short ORFs. For the translated WebLogo consensus sequence in a, amino acids in blue, green, and black are hydrophilic, neutral, and hydrophobic, respectively. The two candidate uORFs in b are associated with shikimate metabolism genes, and notable amino acids related to this pathway and encoded by the uORFs include phenylalanine [F] and tyrosine [Y]. The protein binding candidate in c is depicted with two pyrimidine-rich sequences highlighted that might function as protein binding sites. In addition to the type I consensus depicted, representatives conforming to a type II (only one hairpin-loop similar to P3) and a type III (terminator stem only) consensus also exist. RBS designates ribosome binding sites. Additional annotations are as described in the legend to Fig. 3. f Comprehensive summary of the fate of the unknown IGRs after analysis of the five bacterial genomes examined in this study. Annotations are as described in the legend to Fig. 2c
Similar articles
- Comprehensive discovery of novel structured noncoding RNAs in 26 bacterial genomes.
Brewer KI, Greenlee EB, Higgs G, Yu D, Mirihana Arachchilage G, Chen X, King N, White N, Breaker RR. Brewer KI, et al. RNA Biol. 2021 Dec;18(12):2417-2432. doi: 10.1080/15476286.2021.1917891. Epub 2021 May 10. RNA Biol. 2021. PMID: 33970790 Free PMC article. - The discovery of novel noncoding RNAs in 50 bacterial genomes.
Narunsky A, Higgs GA, Torres BM, Yu D, de Andrade GB, Kavita K, Breaker RR. Narunsky A, et al. Nucleic Acids Res. 2024 May 22;52(9):5152-5165. doi: 10.1093/nar/gkae248. Nucleic Acids Res. 2024. PMID: 38647067 Free PMC article. - Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes.
Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Weinberg Z, et al. Genome Biol. 2010;11(3):R31. doi: 10.1186/gb-2010-11-3-r31. Epub 2010 Mar 15. Genome Biol. 2010. PMID: 20230605 Free PMC article. - Large Noncoding RNAs in Bacteria.
Harris KA, Breaker RR. Harris KA, et al. Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0005-2017. doi: 10.1128/microbiolspec.RWR-0005-2017. Microbiol Spectr. 2018. PMID: 29992899 Free PMC article. Review. - Riboswitch distribution, structure, and function in bacteria.
Pavlova N, Kaloudas D, Penchovsky R. Pavlova N, et al. Gene. 2019 Aug 5;708:38-48. doi: 10.1016/j.gene.2019.05.036. Epub 2019 May 22. Gene. 2019. PMID: 31128223 Review.
Cited by
- Guanidine Biosensors Enable Comparison of Cellular Turn-on Kinetics of Riboswitch-Based Biosensor and Reporter.
Manna S, Truong J, Hammond MC. Manna S, et al. ACS Synth Biol. 2021 Mar 19;10(3):566-578. doi: 10.1021/acssynbio.0c00583. Epub 2021 Mar 1. ACS Synth Biol. 2021. PMID: 33646758 Free PMC article. - The Promising Role of Non-Coding RNAs as Biomarkers and Therapeutic Targets for Leukemia.
Ghazimoradi MH, Karimpour-Fard N, Babashah S. Ghazimoradi MH, et al. Genes (Basel). 2023 Jan 3;14(1):131. doi: 10.3390/genes14010131. Genes (Basel). 2023. PMID: 36672872 Free PMC article. Review. - A second riboswitch class for the enzyme cofactor NAD.
Panchapakesan SSS, Corey L, Malkowski SN, Higgs G, Breaker RR. Panchapakesan SSS, et al. RNA. 2021 Jan;27(1):99-105. doi: 10.1261/rna.077891.120. Epub 2020 Oct 21. RNA. 2021. PMID: 33087526 Free PMC article. - Naturally ornate RNA-only complexes revealed by cryo-EM.
Kretsch RC, Wu Y, Shabalina SA, Lee H, Nye G, Koonin EV, Gao A, Chiu W, Das R. Kretsch RC, et al. Nature. 2025 May 6. doi: 10.1038/s41586-025-09073-0. Online ahead of print. Nature. 2025. PMID: 40328315 - DRAGoM: Classification and Quantification of Noncoding RNA in Metagenomic Data.
Liu B, Thippabhotla S, Zhang J, Zhong C. Liu B, et al. Front Genet. 2021 May 5;12:669495. doi: 10.3389/fgene.2021.669495. eCollection 2021. Front Genet. 2021. PMID: 34025724 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR029676/RR/NCRR NIH HHS/United States
- R01 DE022340/DE/NIDCR NIH HHS/United States
- P01 GM022778/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources