Phasor histone FLIM-FRET microscopy quantifies spatiotemporal rearrangement of chromatin architecture during the DNA damage response - PubMed (original) (raw)
Phasor histone FLIM-FRET microscopy quantifies spatiotemporal rearrangement of chromatin architecture during the DNA damage response
Jieqiong Lou et al. Proc Natl Acad Sci U S A. 2019.
Abstract
To investigate how chromatin architecture is spatiotemporally organized at a double-strand break (DSB) repair locus, we established a biophysical method to quantify chromatin compaction at the nucleosome level during the DNA damage response (DDR). The method is based on phasor image-correlation spectroscopy of histone fluorescence lifetime imaging microscopy (FLIM)-Förster resonance energy transfer (FRET) microscopy data acquired in live cells coexpressing H2B-eGFP and H2B-mCherry. This multiplexed approach generates spatiotemporal maps of nuclear-wide chromatin compaction that, when coupled with laser microirradiation-induced DSBs, quantify the size, stability, and spacing between compact chromatin foci throughout the DDR. Using this technology, we identify that ataxia-telangiectasia mutated (ATM) and RNF8 regulate rapid chromatin decompaction at DSBs and formation of compact chromatin foci surrounding the repair locus. This chromatin architecture serves to demarcate the repair locus from the surrounding nuclear environment and modulate 53BP1 mobility.
Keywords: DNA repair; Förster resonance energy transfer; chromatin organization; fluorescence lifetime imaging microscopy; spatiotemporal correlation spectroscopy.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
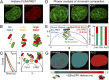
Phasor approach to FLIM-FRET analysis of chromatin compaction. (A) HeLaH2B-2FP nucleus coexpressing H2B-eGFP and H2B-mCherry (H2B-mCh). (B) Graphical depiction of how increasing nucleosome proximity leads to increased FRET between fluorescent histones. (C) Graphical depiction of phasor transformation of HeLaH2B-2FP FLIM-FRET data. (C, Left) Fluorescence lifetime of H2B-eGFP reports on the degree of FRET interaction in each pixel. Each line represents the fluorescent lifetime from a different pixel. (C, Right) These data when phasor-transformed give rise to phasor coordinates (s, g). The donor phasor is right-shifted to shorter fluorescent lifetimes depending on the efficiency of FRET interaction. In HeLaH2B-2FP, decreasing lifetime and increasing FRET corresponds to more compact chromatin. (D) Untreated, TSA-treated, or Actinomycin D (Act D)-treated HeLaH2B-2FP nuclei, shown in the H2B-eGFP channel. (E) Combined phasor distribution of H2B-eGFP fluorescence lifetime from all conditions shown in D with the theoretical FRET trajectory superimposed to determine the range of FRET efficiencies in HeLaH2B-2FP. The linear combination of unquenched donor and background cellular autofluorescence (teal–bright green) (defined in
SI Appendix, Fig. S3
) follows a distinct trajectory from FRET (teal–red). (F) Fraction of pixels in a compact (red) vs. open (teal) chromatin state in control (Cntrl), TSA-, and Act D-treated cells. (G) Pseudocolored chromatin compaction maps of the cells in D according to the palette defined in the phasor plot data in E. (Scale bars, 5 μm.)
Fig. 2.
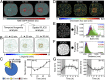
ICS applied to a phasor FLIM-FRET map measures nuclear-wide compact chromatin stability, size, and spacing. (A_–_C) Foci map. (A) FLIM-FRET map from an unirradiated HeLaH2B-2FP nucleus. (B and C) Pixel coordinates of the high FRET state (16–21% FRET) can be extracted from the phasor plot (B) to produce a localization map of compact chromatin (C). (D_–_F) Longevity analysis. (D) Localization map of compacted chromatin from the HeLaH2B-2FP cell in A at 0, 10, and 20 min (as detected by FRET). (E) Schematic of longevity analysis: Averaging three binary images gives rise to a heat map of pixel longevity, which contains structural information that is not evident in the source images and can be used to quantify the stability of detected structures. (F) Longevity map of the compacted chromatin foci detected and tracked in D, with digital enlargement shown for a ROI that contains foci present for 10 min (green pixels) and 20 min (red pixels). The fraction of foci persistence across the time course is calculated as a measure of overall chromatin network stability. (G_–_I) PLICS analysis. (G) A localization map of compacted chromatin from an unirradiated HeLaH2B-2FP nucleus as detected by FLIM-FRET. (H) Schematic of PLICS analysis: In a binary image showing different-sized structures, we can calculate localized 2D spatial correlation functions using an m × m matrix and by collapsing them into 1D correlation profiles. The resulting decay is characteristic of the size of a structure within each m × m matrix. By transforming each decay into phasor coordinates (g, s), we can graphically pseudocolor each pixel according to size. (I) Size map of compacted chromatin foci detected in G and PLICS/iPLICS analysis of the average size (305 nm) and spacing (485 nm). (Scale bars, 5 μm.)
Fig. 3.
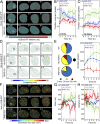
FLIM-FRET analysis of chromatin compaction reveals chromatin architectural changes during the DDR. (A and B) Time series of H2B-eGFP fluorescence intensity images (A) and lifetime maps (B) acquired in a HeLaH2B-2FP 10 min before and at hourly intervals after NIR irradiation. The white square indicates the NIR laser-treated locus. (C) Digital enlargement of the DNA damage site selected in B and the corresponding time series of lifetime maps within this ROI. (D) Masks selected for analysis of the number of pixels in a compacted (high-FRET) vs. noncompacted (low-FRET) state at the DNA damage site vs. outside this ROI. (E and F) Fraction of pixels within (E) and outside (F) of the NIR-irradiated ROI that are in a compacted state during the DDR (red curve) vs. an unperturbed cell (black curve) (n = 10 cells, mean ± SEM). (Scale bars, 5 μm.)
Fig. 4.
A compact chromatin border forms around the lesion site within the first 30 min of the DDR. (A) FLIM-FRET maps acquired in a HeLaH2B-2FP cell during the first 30 min after NIR irradiation. (B) Longevity maps of compact chromatin foci taken at 10-min intervals over a 20-min duration. (C) Quantification of the number of stable compact chromatin pixels across multiple cells (n = 10, mean ± SEM) in the first hour following NIR-induced DSBs (dashed line represents average number of stable compact chromatin pixels in an unperturbed cell). (D) Chromatin compaction size maps derived by PLICS analysis 0–30 min after DSB induction. (E) PLICS analysis of the histogram of sizes induced by DSBs in the data presented in D. (F) iPLICS analysis of the histogram of distance changes induced by DSBs in the data presented in D. (G) Change in mean size and distance between compacted chromatin foci as a function of time during the DDR (n = 10 cells, mean ± SEM). (Scale bars, 5 μm.)
Fig. 5.
ATM and RNF8 regulate chromatin architecture in the DDR. (A) FLIM-FRET maps of HeLaH2B-2FP, HeLaH2B-2FP treated with KU-55933, or HeLaH2B-2FP RNF8 KO cells 10 min before and 1, 3, and 6 h after microirradiation. (B and C) Fraction of compacted chromatin pixels within (B) and outside (C) of the NIR irradiation site in HeLaH2B-2FP (blue curve; n = 10), KU-55933-treated (green curve; n = 6), or RNF8 KO cells (red curve; n = 3; mean ± SEM). (D) Longevity maps of compact chromatin foci from the cells in A. (E) Quantification of compact chromatin foci stability (Left) throughout the DDR (Right) in NIR laser-treated HeLaH2B-2FP, KU-55933–treated HeLaH2B-2FP, and HeLaH2B-2FP RNF8 KO cells (n as indicated above, mean ± SEM). (F) Chromatin compaction size map derived by PLICS from the cells shown in A. (G and H) Change in the mean size of (G) and distance between (H) compacted chromatin foci during the DDR in NIR laser-treated HeLaH2B-2FP, KU-55933–treated HeLaH2B-2FP, and HeLaH2B-2FP RNF8 KO cells (n as indicated above, mean ± SEM). (Scale bars, 5 μm.)
Fig. 6.
Chromatin architecture demarcates the repair locus. (A) FLIM-FRET maps acquired in HeLaH2B-2FP cells 10 min before and 0, 30, and 60 min after microirradiation (Upper) and expanded images of the DSB ROI (Lower). (B) eGFP-53BP1 intensity images acquired in HeLa cells 10 min before and 0, 30, and 60 min after microirradiation (Upper) and expanded images of the DSB ROI (Lower). (C) Correlation of compact chromatin foci localization along the horizontal axis as a function of time (Top; image from 0 min is shown), with 53BP1 localization (Middle; image from 0 min is shown), and mobility in terms of time delay (Bottom). eGFP-53BP1 localization and mobility are averaged along the horizontal axes (green plots). Red dashed box indicates laser microirradiation ROI. (D) Comparison of compact chromatin foci localization (Top), eGFP-53BP1 localization (Middle), and eGFP-53BP1 mobility (Bottom) in untreated HeLa cells (blue curve), KU-55933–treated HeLa cells (green curve), or RNF8 KO cells (red curve) 10 min before and 0, 30, and 60 min after microirradiation. Representative example of n = 3 shown. (Scale bars, 5 μm.)
Similar articles
- Quantifying nuclear wide chromatin compaction by phasor analysis of histone Förster resonance energy transfer (FRET) in frequency domain fluorescence lifetime imaging microscopy (FLIM) data.
Liang Z, Lou J, Scipioni L, Gratton E, Hinde E. Liang Z, et al. Data Brief. 2020 Mar 12;30:105401. doi: 10.1016/j.dib.2020.105401. eCollection 2020 Jun. Data Brief. 2020. PMID: 32300614 Free PMC article. - Phasor Histone FLIM-FRET Microscopy Maps Nuclear-Wide Nanoscale Chromatin Architecture With Respect to Genetically Induced DNA Double-Strand Breaks.
Lou J, Solano A, Liang Z, Hinde E. Lou J, et al. Front Genet. 2021 Dec 10;12:770081. doi: 10.3389/fgene.2021.770081. eCollection 2021. Front Genet. 2021. PMID: 34956323 Free PMC article. - RNF8 promotes high linear energy transfer carbon-ion-induced DNA double-stranded break repair in serum-starved human cells.
Nakajima NI, Yamauchi M, Kakoti S, Cuihua L, Kato R, Permata TBM, Iijima M, Yajima H, Yasuhara T, Yamada S, Hasegawa S, Shibata A. Nakajima NI, et al. DNA Repair (Amst). 2020 Jul-Aug;91-92:102872. doi: 10.1016/j.dnarep.2020.102872. Epub 2020 May 20. DNA Repair (Amst). 2020. PMID: 32502756 - Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization.
Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Costes SV, et al. Mutat Res. 2010 Apr-Jun;704(1-3):78-87. doi: 10.1016/j.mrrev.2009.12.006. Epub 2010 Jan 8. Mutat Res. 2010. PMID: 20060491 Free PMC article. Review. - The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.
Goodarzi AA, Jeggo P, Lobrich M. Goodarzi AA, et al. DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
Cited by
- Direct measurement of protein-protein interactions by FLIM-FRET at UV laser-induced DNA damage sites in living cells.
Kaufmann T, Herbert S, Hackl B, Besold JM, Schramek C, Gotzmann J, Elsayad K, Slade D. Kaufmann T, et al. Nucleic Acids Res. 2020 Dec 2;48(21):e122. doi: 10.1093/nar/gkaa859. Nucleic Acids Res. 2020. PMID: 33053171 Free PMC article. - FLUTE: A Python GUI for interactive phasor analysis of FLIM data.
Gottlieb D, Asadipour B, Kostina P, Ung TPL, Stringari C. Gottlieb D, et al. Biol Imaging. 2023 Nov 6;3:e21. doi: 10.1017/S2633903X23000211. eCollection 2023. Biol Imaging. 2023. PMID: 38487690 Free PMC article. - Histone FRET reports the spatial heterogeneity in nanoscale chromatin architecture that is imparted by the epigenetic landscape at the level of single foci in an intact cell nucleus.
Liang Z, Solano A, Lou J, Hinde E. Liang Z, et al. Chromosoma. 2024 Jan;133(1):5-14. doi: 10.1007/s00412-024-00815-z. Epub 2024 Jan 24. Chromosoma. 2024. PMID: 38265456 Free PMC article. - Tet-Regulated Expression and Optical Clearing for In Vivo Visualization of Genetically Encoded Chimeric dCas9/Fluorescent Protein Probes.
Maloshenok L, Abushinova G, Kazachkina N, Bogdanov A Jr, Zherdeva V. Maloshenok L, et al. Materials (Basel). 2023 Jan 19;16(3):940. doi: 10.3390/ma16030940. Materials (Basel). 2023. PMID: 36769948 Free PMC article. - Phasor S-FLIM: a new paradigm for fast and robust spectral fluorescence lifetime imaging.
Scipioni L, Rossetta A, Tedeschi G, Gratton E. Scipioni L, et al. Nat Methods. 2021 May;18(5):542-550. doi: 10.1038/s41592-021-01108-4. Epub 2021 Apr 15. Nat Methods. 2021. PMID: 33859440 Free PMC article.
References
- Clouaire T, Marnef A, Legube G. Taming tricky DSBs: ATM on duty. DNA Repair (Amst) 2017;56:84–91. - PubMed
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous