Hidden in plain sight: what remains to be discovered in the eukaryotic proteome? - PubMed (original) (raw)
Hidden in plain sight: what remains to be discovered in the eukaryotic proteome?
Valerie Wood et al. Open Biol. 2019.
Abstract
The first decade of genome sequencing stimulated an explosion in the characterization of unknown proteins. More recently, the pace of functional discovery has slowed, leaving around 20% of the proteins even in well-studied model organisms without informative descriptions of their biological roles. Remarkably, many uncharacterized proteins are conserved from yeasts to human, suggesting that they contribute to fundamental biological processes (BP). To fully understand biological systems in health and disease, we need to account for every part of the system. Unstudied proteins thus represent a collective blind spot that limits the progress of both basic and applied biosciences. We use a simple yet powerful metric based on Gene Ontology BP terms to define characterized and uncharacterized proteins for human, budding yeast and fission yeast. We then identify a set of conserved but unstudied proteins in S. pombe, and classify them based on a combination of orthogonal attributes determined by large-scale experimental and comparative methods. Finally, we explore possible reasons why these proteins remain neglected, and propose courses of action to raise their profile and thereby reap the benefits of completing the catalogue of proteins' biological roles.
Keywords: biocuration; budding yeast; fission yeast; gene ontology; human; unknown proteins.
Conflict of interest statement
The authors declare no competing interests.
Figures
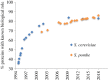
Figure 1.
Characterization history of budding yeast and fission yeast proteins. Numbers of S. pombe and S. cerevisiae proteins that have had their biological roles either determined from experiments or inferred from sequence orthology to known proteins in other species, plotted as a function of time. The numbers of unknown proteins have not markedly decreased over the past 15 years. Data sources: S. cerevisiae 1994–1998 [5], 2000 [6], 2002 [7], 2009 [8], 2013 [9], 2018 this study (figure 3); S. pombe [10].
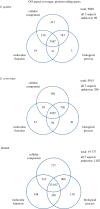
Figure 2.
GO aspect coverage of budding yeast, fission yeast and human proteins. Venn diagrams indicate the number of protein coding gene products annotated to each Gene Ontology aspect (biological process, molecular function, cellular component). Data sources: S. pombe, PomBase 25 September 2018; S. cerevisiae, YeastMine [14] 25 September 2018; human, HumanMine [15] and GO repository [16], both 26 September 2018.
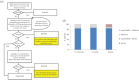
Figure 3.
GO slim analysis of budding yeast, fission yeast and human proteins. (a) Generic GO biological process slim set creation flowchart. The fission yeast GO biological process slim [18] was applied to human and S. cerevisiae protein sets, and then iteratively extended to improve coverage by adding terms. All evidence codes were included except ‘reviewed computational analysis’ (RCA), which yields a higher rate of false positives than the others. Some processes were swapped (e.g. ‘cytoplasmic translation’ in the fission yeast slim for more general ‘translation’) to accommodate the less specific annotation available in other species. The fission yeast slim also omits overly broad terms (e.g. ‘metabolism’) and terms representing activities (molecular functions) in the biological process ontology (e.g. ‘phosphorylation’) because they do not add information about physiological roles; these terms were also excluded from our generic slim set even if inclusion would have increased coverage. (Terms specifically considered but omitted from the generic slim are listed in electronic supplementary material, table S11). At convergence (the point where no additional informative terms could be identified for gene products with biological process annotations), proteins annotated to slim terms were classified as ‘known’ (4393 S. pombe; 4936 S. cerevisiae; 16354 human). The remaining proteins with uninformative processes were classified as unknown, along with those already identified as unknown by annotation to the root node with evidence code ND (no data). Manual assessment of the remaining human proteins with no GO biological process annotation added 266 proteins, bringing the ‘known’ total to 16620. Final ‘unknown’ protein totals are 676 in S. pombe, 978 in S. cerevisiae and 3117 in human. The set of GO slim terms is available in electronic supplementary material, table S1. (b) Proportions of proteins with known GO slim biological role. For all three species, ‘known’ proteins have annotation to at least one term from the GO slim set (see A), and ‘unknown’ proteins do not. Because the human proteome includes some proteins that lack annotation in the GO database, the proportions of unannotated proteins that we found to be known (i.e. annotatable) and unknown are indicated separately. All protein datasets exclude dubious proteins and transposons. Analysis was performed using GOT
erm
F
inder
[21], with GO data from 25 September 2018 and the GO slim created as described in A. Input protein lists are available in electronic supplementary materials, tables S2 (S. pombe), S3 (S. cerevisiae) and S4 (human). GOT
erm
F
inder
output is available in electronic supplementary material, tables S5 (S. pombe), S6 (S. cerevisiae) and S7 (human).
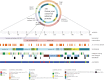
Figure 4.
Taxonomic conservation and features of unknown proteins. Classification of 210 conserved unknown fission yeast proteins along various axes. PomBase curators manually assign protein-coding genes to one of a set of broad taxonomic classifiers [20,29]. PomBase also maintains manually curated lists of orthologues between S. pombe and S. cerevisiae, and between S. pombe and human, three eukaryotic species separated by approximately 500–1000 million years of evolution. In combination, these inventories can be used to identify conservation across taxonomic space at different levels of specificity. Of the fission yeast ‘unknown’ protein-coding genes, 410 are conserved outside the Schizosaccharomyces clade. Of these, 210 are present either in fungi and vertebrates, or in fungi and prokaryotes (data from PomBase manual assignments, queried on 31 July 2018). Proteins were classed as catalytic (i.e. having an identifiable catalytic fold) or non-catalytic (no currently identifiable catalytic fold) based on protein domain, fold, clan or superfamily membership, using InterPro [30] and GO [13] assignments. Cellular locations using GO annotation are available for most of the unknown proteome based on a genome-wide localization study and inference from other models [31]. Viability data come from large-scale screens reported by Kim et al. [32] and Chen et al. [33]. The fission yeast ‘conserved unknown’ protein set [18] is reviewed continually for new functional data.
Similar articles
- The essential function of Rrs1 in ribosome biogenesis is conserved in budding and fission yeasts.
Wan K, Kawara H, Yamamoto T, Kume K, Yabuki Y, Goshima T, Kitamura K, Ueno M, Kanai M, Hirata D, Funato K, Mizuta K. Wan K, et al. Yeast. 2015 Sep;32(9):607-14. doi: 10.1002/yea.3083. Epub 2015 Jul 20. Yeast. 2015. PMID: 26122634 - Genetic approaches to aging in budding and fission yeasts: new connections and new opportunities.
Chen BR, Runge KW. Chen BR, et al. Subcell Biochem. 2012;57:291-314. doi: 10.1007/978-94-007-2561-4_13. Subcell Biochem. 2012. PMID: 22094427 Review. - Proteome effects of genome-wide single gene perturbations.
Öztürk M, Freiwald A, Cartano J, Schmitt R, Dejung M, Luck K, Al-Sady B, Braun S, Levin M, Butter F. Öztürk M, et al. Nat Commun. 2022 Oct 18;13(1):6153. doi: 10.1038/s41467-022-33814-8. Nat Commun. 2022. PMID: 36257942 Free PMC article. - A Proteome-wide Fission Yeast Interactome Reveals Network Evolution Principles from Yeasts to Human.
Vo TV, Das J, Meyer MJ, Cordero NA, Akturk N, Wei X, Fair BJ, Degatano AG, Fragoza R, Liu LG, Matsuyama A, Trickey M, Horibata S, Grimson A, Yamano H, Yoshida M, Roth FP, Pleiss JA, Xia Y, Yu H. Vo TV, et al. Cell. 2016 Jan 14;164(1-2):310-323. doi: 10.1016/j.cell.2015.11.037. Cell. 2016. PMID: 26771498 Free PMC article. - Vesicle-mediated protein transport pathways to the vacuole in Schizosaccharomyces pombe.
Takegawa K, Iwaki T, Fujita Y, Morita T, Hosomi A, Tanaka N. Takegawa K, et al. Cell Struct Funct. 2003 Oct;28(5):399-417. doi: 10.1247/csf.28.399. Cell Struct Funct. 2003. PMID: 14745133 Review.
Cited by
- JaponicusDB: rapid deployment of a model organism database for an emerging model species.
Rutherford KM, Harris MA, Oliferenko S, Wood V. Rutherford KM, et al. Genetics. 2022 Apr 4;220(4):iyab223. doi: 10.1093/genetics/iyab223. Genetics. 2022. PMID: 35380656 Free PMC article. - Functional unknomics: Systematic screening of conserved genes of unknown function.
Rocha JJ, Jayaram SA, Stevens TJ, Muschalik N, Shah RD, Emran S, Robles C, Freeman M, Munro S. Rocha JJ, et al. PLoS Biol. 2023 Aug 8;21(8):e3002222. doi: 10.1371/journal.pbio.3002222. eCollection 2023 Aug. PLoS Biol. 2023. PMID: 37552676 Free PMC article. - From Petri Plates to Petri Nets, a revolution in yeast biology.
Oliver SG. Oliver SG. FEMS Yeast Res. 2022 Feb 22;22(1):foac008. doi: 10.1093/femsyr/foac008. FEMS Yeast Res. 2022. PMID: 35142857 Free PMC article. No abstract available. - Functional repertoire convergence of distantly related eukaryotic plankton lineages abundant in the sunlit ocean.
Delmont TO, Gaia M, Hinsinger DD, Frémont P, Vanni C, Fernandez-Guerra A, Eren AM, Kourlaiev A, d'Agata L, Clayssen Q, Villar E, Labadie K, Cruaud C, Poulain J, Da Silva C, Wessner M, Noel B, Aury JM; Tara Oceans Coordinators; de Vargas C, Bowler C, Karsenti E, Pelletier E, Wincker P, Jaillon O. Delmont TO, et al. Cell Genom. 2022 Apr 28;2(5):100123. doi: 10.1016/j.xgen.2022.100123. eCollection 2022 May 11. Cell Genom. 2022. PMID: 36778897 Free PMC article. - Uncovering the Role of Metabolism in Oomycete-Host Interactions Using Genome-Scale Metabolic Models.
Rodenburg SYA, Seidl MF, de Ridder D, Govers F. Rodenburg SYA, et al. Front Microbiol. 2021 Oct 11;12:748178. doi: 10.3389/fmicb.2021.748178. eCollection 2021. Front Microbiol. 2021. PMID: 34707596 Free PMC article. Review.
References
- NIH. 2018 Genome information by organism. See https://www.ncbi.nlm.nih.gov/genome/browse/ (accessed: 1 October 2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases