Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation - PubMed (original) (raw)
Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation
Duan Chu et al. BMC Cancer. 2019.
Abstract
Background: Nonsynonymous mutations change the protein sequences and are frequently subjected to natural selection. The same goes for nonsense mutations that introduce pre-mature stop codons into CDSs (coding sequences). Synonymous mutations, however, are intuitively thought to be functionally silent and evolutionarily neutral. Now researchers know that the optimized synonymous codon usage is advantageous in the speedy mRNA translation process. With the advent of NGS technique, the explosion of NGS data generated from the tumor tissues help researchers identify driver mutations in cancer-related genes, but relatively less attention is paid to the SNP data in healthy human populations when studying cancer.
Methods: Here, we analyzed the publically available human SNPs. We classified these SNPs according to their functional and evolutionary categories. By simply dividing the human genes into cancer-related genes and other genes, we compared the features of nonsynonymous, synonymous and nonsense mutations in these two gene sets from multiple aspects.
Results: We provided lines of evidence that the nonsynonymous, synonymous and nonsense mutations in cancer-related genes undergo stronger purifying selection when compared to the expected pattern in other genes. The lower nonsynonymous to synonymous ratio observed in cancer-related genes suggests the suppression of amino acid substitutions in these genes. The synonymous SNPs, after excluding those in splicing regions, exhibit preferred changes in codon usage and higher codon frequencies in cancer-related genes compared to other genes, indicating the constraint exerted on these mutations. Nonsense mutations are less frequent and located closer to stop codons in cancer-related genes than in other genes, which putatively minimize their deleterious effects.
Conclusion: Our study demonstrated the evolutionary constraint on mutations in CDS of cancer-related genes without the requirement of data from cancer tissues or patients. Our work provides novel perspectives on interpreting the constraint on mutations in cancer-related genes. We reveal extra constraint on synonymous mutations in cancer-related genes which is related to codon usage bias and is in addition to the splicing effect.
Keywords: Cancer-related genes; Coding region; Codon usage bias (CUB); Mutation; Natural selection.
Conflict of interest statement
Ethics approval and consent to participate
All datasets used in this study were downloaded from publically available websites as described in the Methods section.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
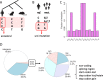
The landscape of human SNPs. a A schematic diagram showing the filtering of SNPs according to the orthologous sites in rhesus monkey and mouse genomes. The phylogenetic tree is unscaled. b A schematic diagram displaying the definition of uni-mutation SNPs. ref., reference. Mut., mutation. c the fractions of different types of mutations. d The annotations and proportions of genomic SNPs (left) and exonic SNPs (right) after the above filtering steps. The clip arts of human/monkey/mouse were drawn by ourselves
Fig. 2
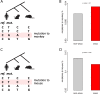
Constraint on mutations belonging to different evolutionary categories. a A schematic diagram showing the definition of mutation to monkey. The phylogenetic tree is unscaled. b Fractions of SNPs that the reference allele is observed in mouse and the alternative allele is observed in monkey. _P_-value was calculated using Fisher’s exact test. c A schematic diagram showing the definition of mutation to mouse. The phylogenetic tree is unscaled. d Fractions of SNPs that the reference allele is observed in monkey and the alternative allele is observed in mouse. _P_-value was calculated using Fisher’s exact test. “onco” denotes cancer-related genes; “non-onco” denotes other genes. The clip arts of human/monkey/mouse were drawn by ourselves
Fig. 3
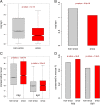
Purifying selection on the nonsynonymous as well as the synonymous SNPs in cancer-related genes. a Comparison between the nonsynonymous to synonymous ratio (nsy/syn) of cancer-related genes and other genes. _P_-value was calculated using Wilcoxon rank sum test. b Comparison between the pooled nsy/syn ratio of cancer-related genes and other genes. P-value was calculated using Fisher’s exact test. c Conservation level (phyloP score) of nonsynonymous and synonymous SNPs in cancer-related genes and other genes. _P_-values were calculated using Wilcoxon rank sum tests. d Fractions of conserved orthologous sites in mouse genome. _P_-values were calculated using Fisher’s exact tests. “onco” denotes cancer-related genes; “non-onco” denotes other genes
Fig. 4
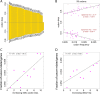
Purifying selection on the synonymous SNPs in cancer-related genes. a Codon bias (codon preference) of the 59 sense codons in human genome. Codons with a positive value are preferred, and vice versa. b Correlation between codon bias and codon frequency appearing in human genome. c Correlation between the delta codon bias after the synonymous change and the fraction of cancer-related genes. Genes were divided into ten bins with increasing delta codon bias. d Correlation between the delta codon frequency after the synonymous change and the fraction of cancer-related genes. Genes were divided into ten bins with increasing delta codon frequency. “onco” denotes cancer-related genes; “non-onco” denotes other genes
Fig. 5
Purifying selection on the nonsense mutations that create stop codons in coding region. a Relative position of the nonsense variants in cancer-related genes and other genes. _P_-value was calculated using Wilcoxon rank sum test. b Comparison between the nonsense/syn ratios in cancer-related genes and other genes. P-value was calculated using Fisher’s exact test. c A diagram illustrating the effects of double-mutation within a codon. Quite a lot of double-mutations result in a stop codon in CDS. d Comparison between the double-mutation/syn ratios in cancer-related genes and other genes. _P_-value was calculated using Fisher’s exact test. e Relative position of the double-mutations that create stop codons cancer-related genes and other genes. “onco” denotes cancer-related genes; “non-onco” denotes other genes
Similar articles
- Parsing the synonymous mutations in the maize genome: isoaccepting mutations are more advantageous in regions with codon co-occurrence bias.
Chu D, Wei L. Chu D, et al. BMC Plant Biol. 2019 Oct 14;19(1):422. doi: 10.1186/s12870-019-2050-1. BMC Plant Biol. 2019. PMID: 31610786 Free PMC article. - Natural selection pressure exerted on "Silent" mutations during the evolution of SARS-CoV-2: Evidence from codon usage and RNA structure.
Bai H, Ata G, Sun Q, Rahman SU, Tao S. Bai H, et al. Virus Res. 2023 Jan 2;323:198966. doi: 10.1016/j.virusres.2022.198966. Epub 2022 Oct 14. Virus Res. 2023. PMID: 36244617 Free PMC article. - Codon-Resolution Analysis Reveals a Direct and Context-Dependent Impact of Individual Synonymous Mutations on mRNA Level.
Chen S, Li K, Cao W, Wang J, Zhao T, Huan Q, Yang YF, Wu S, Qian W. Chen S, et al. Mol Biol Evol. 2017 Nov 1;34(11):2944-2958. doi: 10.1093/molbev/msx229. Mol Biol Evol. 2017. PMID: 28961875 Free PMC article. - Effects of Synonymous Mutations beyond Codon Bias: The Evidence for Adaptive Synonymous Substitutions from Microbial Evolution Experiments.
Bailey SF, Alonso Morales LA, Kassen R. Bailey SF, et al. Genome Biol Evol. 2021 Sep 1;13(9):evab141. doi: 10.1093/gbe/evab141. Genome Biol Evol. 2021. PMID: 34132772 Free PMC article. Review. - The Code of Silence: Widespread Associations Between Synonymous Codon Biases and Gene Function.
Supek F. Supek F. J Mol Evol. 2016 Jan;82(1):65-73. doi: 10.1007/s00239-015-9714-8. Epub 2015 Nov 4. J Mol Evol. 2016. PMID: 26538122 Review.
Cited by
- Codon Usage is Influenced by Compositional Constraints in Genes Associated with Dementia.
Alqahtani T, Khandia R, Puranik N, Alqahtani AM, Alghazwani Y, Alshehri SA, Chidambaram K, Kamal MA. Alqahtani T, et al. Front Genet. 2022 Aug 9;13:884348. doi: 10.3389/fgene.2022.884348. eCollection 2022. Front Genet. 2022. PMID: 36017501 Free PMC article. - Mutation Load in Sunflower Inversions Is Negatively Correlated with Inversion Heterozygosity.
Huang K, Ostevik KL, Elphinstone C, Todesco M, Bercovich N, Owens GL, Rieseberg LH. Huang K, et al. Mol Biol Evol. 2022 May 3;39(5):msac101. doi: 10.1093/molbev/msac101. Mol Biol Evol. 2022. PMID: 35535689 Free PMC article. - Synonymous mutations that regulate translation speed might play a non-negligible role in liver cancer development.
Li Q, Li J, Yu CP, Chang S, Xie LL, Wang S. Li Q, et al. BMC Cancer. 2021 Apr 9;21(1):388. doi: 10.1186/s12885-021-08131-w. BMC Cancer. 2021. PMID: 33836673 Free PMC article. - Genetic Polymorphisms of IGF1 and IGF1R Genes and Their Effects on Growth Traits in Hulun Buir Sheep.
Ding N, Tian D, Li X, Zhang Z, Tian F, Liu S, Han B, Liu D, Zhao K. Ding N, et al. Genes (Basel). 2022 Apr 9;13(4):666. doi: 10.3390/genes13040666. Genes (Basel). 2022. PMID: 35456472 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources