Asgard archaea capable of anaerobic hydrocarbon cycling - PubMed (original) (raw)
Asgard archaea capable of anaerobic hydrocarbon cycling
Kiley W Seitz et al. Nat Commun. 2019.
Abstract
Large reservoirs of natural gas in the oceanic subsurface sustain complex communities of anaerobic microbes, including archaeal lineages with potential to mediate oxidation of hydrocarbons such as methane and butane. Here we describe a previously unknown archaeal phylum, Helarchaeota, belonging to the Asgard superphylum and with the potential for hydrocarbon oxidation. We reconstruct Helarchaeota genomes from metagenomic data derived from hydrothermal deep-sea sediments in the hydrocarbon-rich Guaymas Basin. The genomes encode methyl-CoM reductase-like enzymes that are similar to those found in butane-oxidizing archaea, as well as several enzymes potentially involved in alkyl-CoA oxidation and the Wood-Ljungdahl pathway. We suggest that members of the Helarchaeota have the potential to activate and subsequently anaerobically oxidize hydrothermally generated short-chain hydrocarbons.
Conflict of interest statement
The authors declare no competing interests.
Figures
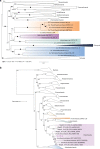
Fig. 1
Phylogenomic position of Helarchaeota within the Asgard archaea superphylum. a Phylogenomic analysis of 56 concatenated ribosomal proteins identified in Helarchaeota bins. Black circles indicate bootstrap values greater than 95 (LG+C60+F+G+PMSF); posterior probability ≥ 0.95 (SR4). b Maximum-likelihood phylogenetic tree of 16S rRNA gene sequences thought to belong to Helarchaeota. The phylogeny was generated using RAxML (GTRGAMMA model and number of bootstraps determined using the extended majority-rule consensus tree criterion). The purple box shows possible Helarchaeota sequences from GB data, as well as closely related published sequences and sequences form recently identified Helarchaeota bins (identified as Megxx_xxxx_Bin_xxx_scaffold_xxxxx). Number of sequences is depicted in the collapsed clades
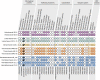
Fig. 2
Distribution of eukaryotic signature proteins (ESPs) in Helarchaeota and other Asgard archaea. Numbers under each column correspond to the InterPro accession number (IPR) and Archaeal Clusters of Orthologous Genes (arcCOG) IDs that were searched for. Full circles refer to cases in which a homolog was found in the respective genomes. Empty circles with black outlines represent the absence of the ESP. The checkered pattern in the RNA polymerase subunit alpha represents the fact that the proteins were split, while the fused proteins are represented by the full circles. Gray circles with borders in any other color represent cases where the standard profiles were not found but potential homologs where detected. In the Roadblock proteins, potential homologs were detected, but the phylogeny could not support the close relationship of any of these copies to the Asgard archaea group closest to eukaryotes. In the Ub-activating enzyme E1 represents homologs found clustered appropriately with its potential orthologs in the phylogeny but the synteny of this gene with other ubiquitin-related proteins in the genome is uncertain
Fig. 3
Metabolic inference of Helarchaeota and phylogenetic analyses of McrAB proteins. a Enzymes shown in dark purple are present in both genomes, those shown in light purple are present in a single genome and ones in gray are absent. b The tree of concatenated McrAB proteins was generated using IQ-tree with 1000 ultrafast bootstraps, single-branch test bootstraps and posterior probability values from the Bayesian phylogeny. White circles indicate support values of 90–99.9/90–99.9/0.9–0.99 and black filled circles indicate values of 100/100/1. The tree was rooted arbitrarily between the cluster comprising canonical McrAB homologs and divergent McrAB homologs. Scale bars indicate the average number of substitutions per site
Fig. 4
Comparison of Helarchaeota metabolism to other alkane oxidizing and methanogenic archaea. Alkane metabolism of Helarchaeota compared to Bathyarchaeota and Ca. Syntrophoarchaeum sp., Verstraetearchaeota, GoM-Arc1 sp., ANME-1 sp., and ANME-2 sp. A list of genes and corresponding contig identifiers can be found in Supplementary Data 1. Filled circles represent the presences of genes and white circles represents absence of genes
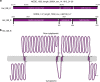
Fig. 5
Gene cluster found in Helarchaeota that encode for a possible energy-transferring complex. a In Hel_GB_A the complex was found on the reverse strand but has been oriented in the forward direction for clarity (asterisk). Arrows indicate the length of the reading frame. Gene names were predicted by various databases (Supplementary Methods). Small numbers located above the arrows refer to the nucleotide position for the full contig. Bold numbers on Hel_GB_B refer to the amino acid number of the whole complex. b Figure depicts the membrane motifs identified on NODE_147_length_7209_cov_4.62199_5, 6, and 7 using various programs (Supplementary Methods). Each circle represents a single amino acid. Bold circles represent amino acids at the start of the protein, the start and end of the transmembrane sites, and the end of the complex. Numbering corresponds to the amino acid numbers of Hel_GB_B in panel a. A full loop represents 50 amino acids and does not reflect the secondary structure of the complex
Similar articles
- Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments.
Dombrowski N, Seitz KW, Teske AP, Baker BJ. Dombrowski N, et al. Microbiome. 2017 Aug 23;5(1):106. doi: 10.1186/s40168-017-0322-2. Microbiome. 2017. PMID: 28835260 Free PMC article. - Brockarchaeota, a novel archaeal phylum with unique and versatile carbon cycling pathways.
De Anda V, Chen LX, Dombrowski N, Hua ZS, Jiang HC, Banfield JF, Li WJ, Baker BJ. De Anda V, et al. Nat Commun. 2021 Apr 23;12(1):2404. doi: 10.1038/s41467-021-22736-6. Nat Commun. 2021. PMID: 33893309 Free PMC article. - "Candidatus Ethanoperedens," a Thermophilic Genus of Archaea Mediating the Anaerobic Oxidation of Ethane.
Hahn CJ, Laso-Pérez R, Vulcano F, Vaziourakis KM, Stokke R, Steen IH, Teske A, Boetius A, Liebeke M, Amann R, Knittel K, Wegener G. Hahn CJ, et al. mBio. 2020 Apr 21;11(2):e00600-20. doi: 10.1128/mBio.00600-20. mBio. 2020. PMID: 32317322 Free PMC article. - Beyond methane, new frontiers in anaerobic microbial hydrocarbon utilizing pathways.
Sarno N, Hyde E, De Anda V, Baker BJ. Sarno N, et al. Microb Biotechnol. 2024 Jun;17(6):e14508. doi: 10.1111/1751-7915.14508. Microb Biotechnol. 2024. PMID: 38888492 Free PMC article. Review. - Methyl/alkyl-coenzyme M reductase-based anaerobic alkane oxidation in archaea.
Wang Y, Wegener G, Ruff SE, Wang F. Wang Y, et al. Environ Microbiol. 2021 Feb;23(2):530-541. doi: 10.1111/1462-2920.15057. Epub 2020 May 18. Environ Microbiol. 2021. PMID: 32367670 Review.
Cited by
- Unraveling the Metabolic Potential of Asgardarchaeota in a Sediment from the Mediterranean Hydrocarbon-Contaminated Water Basin Mar Piccolo (Taranto, Italy).
Firrincieli A, Negroni A, Zanaroli G, Cappelletti M. Firrincieli A, et al. Microorganisms. 2021 Apr 16;9(4):859. doi: 10.3390/microorganisms9040859. Microorganisms. 2021. PMID: 33923677 Free PMC article. - Anaerobic hydrocarbon biodegradation by alkylotrophic methanogens in deep oil reservoirs.
Zhang CJ, Zhou Z, Cha G, Li L, Fu L, Liu LY, Yang L, Wegener G, Cheng L, Li M. Zhang CJ, et al. ISME J. 2024 Jan 8;18(1):wrae152. doi: 10.1093/ismejo/wrae152. ISME J. 2024. PMID: 39083033 Free PMC article. - Anaerobic hexadecane degradation by a thermophilic Hadarchaeon from Guaymas Basin.
Benito Merino D, Lipp JS, Borrel G, Boetius A, Wegener G. Benito Merino D, et al. ISME J. 2024 Jan 8;18(1):wrad004. doi: 10.1093/ismejo/wrad004. ISME J. 2024. PMID: 38365230 Free PMC article. - Metagenomes from Coastal Marine Sediments Give Insights into the Ecological Role and Cellular Features of _Loki_- and Thorarchaeota.
Manoharan L, Kozlowski JA, Murdoch RW, Löffler FE, Sousa FL, Schleper C. Manoharan L, et al. mBio. 2019 Sep 10;10(5):e02039-19. doi: 10.1128/mBio.02039-19. mBio. 2019. PMID: 31506313 Free PMC article. - Well-hidden methanogenesis in deep, organic-rich sediments of Guaymas Basin.
Bojanova DP, De Anda VY, Haghnegahdar MA, Teske AP, Ash JL, Young ED, Baker BJ, LaRowe DE, Amend JP. Bojanova DP, et al. ISME J. 2023 Nov;17(11):1828-1838. doi: 10.1038/s41396-023-01485-y. Epub 2023 Aug 18. ISME J. 2023. PMID: 37596411 Free PMC article.
References
- Claypool GE, Kvenvolden, K. A. Methane and other hydrocarbon gases in marine sediment. Annu. Rev. Earth Planet. Sci. 1983;11:299–327. doi: 10.1146/annurev.ea.11.050183.001503. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources