The evolutionary landscape of the Rab family in chordates - PubMed (original) (raw)
The evolutionary landscape of the Rab family in chordates
Ugo Coppola et al. Cell Mol Life Sci. 2019 Oct.
Abstract
Intracellular traffic amongst organelles represents a key feature for eukaryotes and is orchestrated principally by members of Rab family, the largest within Ras superfamily. Given that variations in Rab repertoire have been fundamental in animal diversification, we provided the most exhaustive survey regarding the Rab toolkit of chordates. Our findings reveal the existence of 42 metazoan conserved subfamilies exhibiting a univocal intron/exon structure preserved from cnidarians to vertebrates. Since the current view does not capture the Rab complexity, we propose a new Rab family classification in three distinct monophyletic clades. The Rab complement of chordates shows a dramatic diversification due to genome duplications and independent gene duplications and losses with sharp differences amongst cephalochordates, tunicates and gnathostome vertebrates. Strikingly, the analysis of the domain architecture of this family highlighted the existence of chimeric calcium-binding Rabs, which are animal novelties characterized by a complex evolutionary history in gnathostomes and whose role in cellular metabolism is obscure. This work provides novel insights in the knowledge of Rab family: our hypothesis is that chordates represent a hotspot of Rab variability, with many events of gene gains and losses impacting intracellular traffic capabilities. Our results help to elucidate the role of Rab members in the transport amongst endomembranes and shed light on intracellular traffic routes in vertebrates. Then, since the predominant role of Rabs in the molecular communication between different cellular districts, this study paves to way to comprehend inherited or acquired human disorders provoked by dysfunctions in Rab genes.
Keywords: Amphioxus Branchiostoma; Ascidian Ciona; Calcium-binding Rab chimeras; Larvacean Oikopleura; Metazoan Rabs; Small GTPase superfamily.
Figures
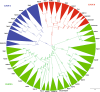
Fig. 1
Rab phylogeny in Metazoa. The cladogram shows three monophyletic Rab clades highlighted with different colours (A green, B red and C blue) encompassing Rab proteins of Nematostella vectensis, Caenorhabditis elegans, Capitella teleta, Lottia gigantea, Saccoglossus kowalevskii, Strongylocentrotus purpuratus, Branchiostoma lanceolatum, Ciona robusta, Oikopleura dioica, Anolis carolinensis, Homo sapiens. Values at the branches represent replicates obtained using aLRT and aBayes methods
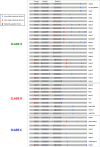
Fig. 2
Rab toolkit: gene duplications and losses shaped the extant metazoan Rab complement. The number of Rab members for each species is indicated in correspondence of the column (#); red numbers represent Rab genes considered to be LECA, according to the classification proposed by Elias et al. [22], plus the Ran gene and without RabTitan and Rab50. The colour of dots indicates the gene presence (black), gene absence (white), invertebrate lineage-specific duplications (orange), vertebrate whole-genome duplicates (green), vertebrate-specific gene duplicates (blue), reptile-specific duplicates (brown), mammalian-specific duplicates (magenta) and primate-specific duplicates (yellow)
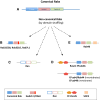
Fig. 3
Rab intron code. Schematization of metazoan Rab gene structure showing the intron conservation code specific for each subfamily. Grey boxes represent the three canonical Rab domains: P-loop, Switch I and Switch II. The green, red and blue frames correspond to the three monophyletic clades resulted from the phylogenetic analysis. The intron/exon boundary of each Rab subfamily is shown by small vertical bars, in blue for intra-clade and in red for inter-clade conserved intron positions
Fig. 4
Modular domain organization. Distinct Rab domain architectures found in metazoans. a Most of Rabs contain “canonical” Rab domains made of P-Loop (bluebox), Switch I (pink box) and Switch II (green box). b The FALK stretch (in red) is exclusive of Rab32, Rab38, Rab32LO and Rab7L1. c Ran proteins possess a divergent amino acid composition in P-Loop, Switch I and Switch II domains (empty boxes). d One or two EF-Hand domains at N-terminus (orange and red pentagons) characterize the chimeras Rab (Rasef, EFcab4/Rab44, EFcab4, and Rab44). e The unique case of Rab40 protein, which contains an SOCS box at the C terminus (light orange box)
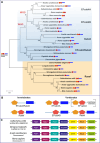
Fig. 5
Evolutionary history of Rab chimeras. a Phylogenetic three of Rab chimeras. EFcab4/Rab44 already present in invertebrates underwent two rounds of genome duplication in vertebrates generating Rab44, EFcab4A and Efcab4B. On the other hand, Rasef is a single-copy gene in both invertebrates and gnathostomes; Rab46 was excluded for their sequence divergence. First and second EF-Hand domains (orange and red pentagons, respectively) and Rab motifs (blue bar) are depicted. On the left, the ancestral organization of Rab chimeras is shown. Numbers at the branches are replicates obtained employing the ML estimation method. b Domain organization of invertebrate EFcab4/Rab44 and gnathostome EFcab4A and EFcab4B. EFcab4A in amphibians and mammals has lost the canonical Rab domain. Rab44 was excluded here for their variability in domain organization. c The orthology of gnathostome EFcab4A genes is demonstrated by the conserved neighbourhood on respective chromosomes
Similar articles
- Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis.
Klöpper TH, Kienle N, Fasshauer D, Munro S. Klöpper TH, et al. BMC Biol. 2012 Aug 8;10:71. doi: 10.1186/1741-7007-10-71. BMC Biol. 2012. PMID: 22873208 Free PMC article. - Rab32 and Rab38 genes in chordate pigmentation: an evolutionary perspective.
Coppola U, Annona G, D'Aniello S, Ristoratore F. Coppola U, et al. BMC Evol Biol. 2016 Jan 27;16:26. doi: 10.1186/s12862-016-0596-1. BMC Evol Biol. 2016. PMID: 26818140 Free PMC article. - Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila.
Bright LJ, Kambesis N, Nelson SB, Jeong B, Turkewitz AP. Bright LJ, et al. PLoS Genet. 2010 Oct 14;6(10):e1001155. doi: 10.1371/journal.pgen.1001155. PLoS Genet. 2010. PMID: 20976245 Free PMC article. - How much does the amphioxus genome represent the ancestor of chordates?
Louis A, Roest Crollius H, Robinson-Rechavi M. Louis A, et al. Brief Funct Genomics. 2012 Mar;11(2):89-95. doi: 10.1093/bfgp/els003. Epub 2012 Feb 28. Brief Funct Genomics. 2012. PMID: 22373648 Free PMC article. Review. - Enhancer evolution in chordates: Lessons from functional analyses of cephalochordate cis-regulatory modules.
Yasuoka Y. Yasuoka Y. Dev Growth Differ. 2020 Jun;62(5):279-300. doi: 10.1111/dgd.12684. Epub 2020 Jun 16. Dev Growth Differ. 2020. PMID: 32479656 Review.
Cited by
- The interrelated roles of RAB family proteins in the advancement of neoplastic growth.
Ji Y, Li R, Tang G, Wang W, Chen C, Yang Q. Ji Y, et al. Front Oncol. 2025 Mar 24;15:1513360. doi: 10.3389/fonc.2025.1513360. eCollection 2025. Front Oncol. 2025. PMID: 40196733 Free PMC article. Review. - Gene expression and cellular changes in injured myocardium of Ciona intestinalis.
Stokes S, Palmer PP, Barth JL, Price RL, Parker BG, Evans Anderson HJ. Stokes S, et al. Front Cell Dev Biol. 2024 Mar 13;12:1304755. doi: 10.3389/fcell.2024.1304755. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38544819 Free PMC article. - The _Cis_-Regulatory Code for Kelch-like 21/30 Specific Expression in Ciona robusta Sensory Organs.
Coppola U, Kamal AK, Stolfi A, Ristoratore F. Coppola U, et al. Front Cell Dev Biol. 2020 Sep 11;8:569601. doi: 10.3389/fcell.2020.569601. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33043001 Free PMC article. - Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis.
Zhang W, Zhang Z, Xiang Y, Gu DD, Chen J, Chen Y, Zhai S, Liu Y, Jiang T, Liu C, He B, Yan M, Wang Z, Xu J, Cao YL, Deng B, Zeng D, Lei J, Zhuo J, Lei X, Long Z, Jin B, Chen T, Li D, Shen Y, Hu J, Gao S, Liu Q. Zhang W, et al. Nat Struct Mol Biol. 2024 Feb;31(2):219-231. doi: 10.1038/s41594-023-01165-7. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177680 - Gene expression in notochord and nuclei pulposi: a study of gene families across the chordate phylum.
Raghavan R, Coppola U, Wu Y, Ihewulezi C, Negrón-Piñeiro LJ, Maguire JE, Hong J, Cunningham M, Kim HJ, Albert TJ, Ali AM, Saint-Jeannet JP, Ristoratore F, Dahia CL, Di Gregorio A. Raghavan R, et al. BMC Ecol Evol. 2023 Oct 27;23(1):63. doi: 10.1186/s12862-023-02167-1. BMC Ecol Evol. 2023. PMID: 37891482 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources