Bioinformatic Mapping of Radical S-Adenosylmethionine-Dependent Ribosomally Synthesized and Post-Translationally Modified Peptides Identifies New Cα, Cβ, and Cγ-Linked Thioether-Containing Peptides - PubMed (original) (raw)
Bioinformatic Mapping of Radical S-Adenosylmethionine-Dependent Ribosomally Synthesized and Post-Translationally Modified Peptides Identifies New Cα, Cβ, and Cγ-Linked Thioether-Containing Peptides
Graham A Hudson et al. J Am Chem Soc. 2019.
Erratum in
- Correction to "Bioinformatic Mapping of Radical _S_-Adenosylmethionine-Dependent Ribosomally Synthesized and Post-Translationally Modified Peptides Identifies New Cα, Cβ, and Cγ-Linked Thioether-Containing Peptides".
Hudson GA, Burkhart BJ, DiCaprio AJ, Schwalen CJ, Kille B, Pogorelov TV, Mitchell DA. Hudson GA, et al. J Am Chem Soc. 2021 Jul 14;143(27):10479. doi: 10.1021/jacs.1c06285. Epub 2021 Jul 1. J Am Chem Soc. 2021. PMID: 34197098 No abstract available.
Abstract
Recently developed bioinformatic tools have bolstered the discovery of ribosomally synthesized and post-translationally modified peptides (RiPPs). Using an improved version of Rapid ORF Description and Evaluation Online (RODEO 2.0), a biosynthetic gene cluster mining algorithm, we bioinformatically mapped the sactipeptide RiPP class via the radical S-adenosylmethionine (SAM) enzymes that form the characteristic sactionine (sulfur-to-α carbon) cross-links between cysteine and acceptor residues. Hundreds of new sactipeptide biosynthetic gene clusters were uncovered, and a novel sactipeptide "huazacin" with growth-suppressive activity against Listeria monocytogenes was characterized. Bioinformatic analysis further suggested that a group of sactipeptide-like peptides heretofore referred to as six cysteines in forty-five residues (SCIFFs) might not be sactipeptides as previously thought. Indeed, the bioinformatically identified SCIFF peptide "freyrasin" was demonstrated to contain six thioethers linking the β carbons of six aspartate residues. Another SCIFF, thermocellin, was shown to contain a thioether cross-linked to the γ carbon of threonine. SCIFFs feature a different paradigm of non-α carbon thioether linkages, and they are exclusively formed by radical SAM enzymes, as opposed to the polar chemistry employed during lanthipeptide biosynthesis. Therefore, we propose the renaming of the SCIFF family as radical non-α thioether peptides (ranthipeptides) to better distinguish them from the sactipeptide and lanthipeptide RiPP classes.
Figures
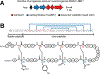
Figure 1.. Sactipeptide biosynthesis and ring topologies.
(A) Schematic overview of sactipeptide biosynthesis. (B) Proposed mechanism for the rSAM-dependent formation of the sactipeptide S–Cα thioether crosslink. (C) Precursor peptide sequence and ring topologies of characterized sactipeptides (blue, S–Cα linkage; yellow, S-S linkage; black, uncharacterized). The requisite rSAM enzyme name is given in parentheses. Characterization of the six Cys in forty-five residues (SCIFFs) is limited to in vitro enzymatic assay for which only one thioether linkage was observed. Data shown later in this manuscript support a Cys-Thr linkage different to what has been previously reported and further that SCIFFs are not sactipeptides. * Hyicin 4244 has not been structurally characterized, thus the linkages indicated are speculative.
Figure 2.. Huazacin from Bacillus thuringiensis serovar huazhongensis.
(A) Biosynthetic gene cluster. The sactipeptide synthases HuaB and HuaC (NCBI accession identifiers: EEM79976.1 and EEM79977.1, respectively) feature an N-terminal RRE domain and C-terminal SPASM domain similar to other sactisynthases., (B) Precursor peptide sequence and chemical structure of huazacin. The hua gene cluster contains two identical copies of the precursor peptide shown.
Figure 3.. Sequence similarity network of rSAM enzymes.
rSAM enzymes involved in sactipeptide and SCIFF biosynthesis were used as BLAST inputs to gather additional rSAM enzymes predicted to form RiPP thioether linkages (n = 4,693). Sequences were analyzed by EFI-EST and visualized using Cytoscape. Nodes are colored based on the category of its associated precursor peptide (see legend). Sequences sharing >70% identity are conflated as a single node, and connected nodes indicate a similarity score of < 1e-50.,, Sequence logos for core peptides associated with these rSAMs are shown in Figures S1 and S9. QhpD, non-RiPP rSAM involved in modifying quinohemoprotein amine dehydrogenase.
Figure 4.. Freyrasin biosynthetic gene cluster and related peptides.
(A) Biosynthetic gene cluster diagram. (B) Freyrasin precursor peptide sequence with six CX3D motifs and thioether topologies highlighted. (C) Alignment of additional freyrasin members. Leader peptide residues are negatively numbered relative to the core peptide, which we predict begins with Gly.
Figure 5.. Freyrasin contains beta-linked thioethers.
(A) Cys-Asp (S–Cβ) connectivity with hydrogens colored pink (Cys) or blue (Asp). (B) 1H-1H NOESY correlations confirming a S–Cβ thioether linkage between Cys26-Asp30. (C) Same as panel B but for Cys20-Asp24.
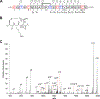
Figure 6.. Thermocellin contains a Cys-Thr (S-Cγ) thioether.
(A) Coexpression of the thermocellin precursor containing labeled Thr (blue) features a −2 Da shift relative to unmodified peptide, demonstrating a S-Cγ linkage was formed. The modified peptide was treated with endoproteinase GluC and unmodified Cys were alkylated with _N_-ethylmaleimide (red) Residues are numbered based on the sequence after GluC treatment. (B) Structure of the isotopically labeled thioether linkage. (C) MS/MS analysis yielded fragmentation consistent with a macrocycle formed between Cys9 and Thr11. Error tables for assigned ions are available in Table S6.
Similar articles
- Reconstitution and Substrate Specificity of the Thioether-Forming Radical _S_-Adenosylmethionine Enzyme in Freyrasin Biosynthesis.
Precord TW, Mahanta N, Mitchell DA. Precord TW, et al. ACS Chem Biol. 2019 Sep 20;14(9):1981-1989. doi: 10.1021/acschembio.9b00457. Epub 2019 Sep 9. ACS Chem Biol. 2019. PMID: 31449382 Free PMC article. - Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
Grove TL, Himes PM, Hwang S, Yumerefendi H, Bonanno JB, Kuhlman B, Almo SC, Bowers AA. Grove TL, et al. J Am Chem Soc. 2017 Aug 30;139(34):11734-11744. doi: 10.1021/jacs.7b01283. Epub 2017 Aug 21. J Am Chem Soc. 2017. PMID: 28704043 Free PMC article. - SkfB Abstracts a Hydrogen Atom from Cα on SkfA To Initiate Thioether Cross-Link Formation.
Bruender NA, Bandarian V. Bruender NA, et al. Biochemistry. 2016 Aug 2;55(30):4131-4. doi: 10.1021/acs.biochem.6b00598. Epub 2016 Jul 21. Biochemistry. 2016. PMID: 27410522 Free PMC article. - Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis.
Flühe L, Marahiel MA. Flühe L, et al. Curr Opin Chem Biol. 2013 Aug;17(4):605-12. doi: 10.1016/j.cbpa.2013.06.031. Epub 2013 Jul 24. Curr Opin Chem Biol. 2013. PMID: 23891473 Review. - Current Advancements in Sactipeptide Natural Products.
Chen Y, Wang J, Li G, Yang Y, Ding W. Chen Y, et al. Front Chem. 2021 May 20;9:595991. doi: 10.3389/fchem.2021.595991. eCollection 2021. Front Chem. 2021. PMID: 34095082 Free PMC article. Review.
Cited by
- Cutting the Gordian knot: early and complete amino acid sequence confirmation of class II lasso peptides by HCD fragmentation.
Jarmusch SA, Feldmann I, Blank-Landeshammer B, Cortés-Albayay C, Castro JF, Andrews B, Asenjo JA, Sickmann A, Ebel R, Jaspars M. Jarmusch SA, et al. J Antibiot (Tokyo). 2020 Nov;73(11):772-779. doi: 10.1038/s41429-020-00369-z. Epub 2020 Sep 9. J Antibiot (Tokyo). 2020. PMID: 32908238 - Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides.
Ongpipattanakul C, Desormeaux EK, DiCaprio A, van der Donk WA, Mitchell DA, Nair SK. Ongpipattanakul C, et al. Chem Rev. 2022 Sep 28;122(18):14722-14814. doi: 10.1021/acs.chemrev.2c00210. Epub 2022 Sep 1. Chem Rev. 2022. PMID: 36049139 Free PMC article. Review. - Emulating nonribosomal peptides with ribosomal biosynthetic strategies.
Mordhorst S, Ruijne F, Vagstad AL, Kuipers OP, Piel J. Mordhorst S, et al. RSC Chem Biol. 2022 Dec 6;4(1):7-36. doi: 10.1039/d2cb00169a. eCollection 2023 Jan 4. RSC Chem Biol. 2022. PMID: 36685251 Free PMC article. Review. - ThnL, a B12-dependent radical _S_-adenosylmethionine enzyme, catalyzes thioether bond formation in carbapenem biosynthesis.
Sinner EK, Li R, Marous DR, Townsend CA. Sinner EK, et al. Proc Natl Acad Sci U S A. 2022 Aug 23;119(34):e2206494119. doi: 10.1073/pnas.2206494119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969793 Free PMC article. - Translation-Targeting RiPPs and Where to Find Them.
Travin DY, Bikmetov D, Severinov K. Travin DY, et al. Front Genet. 2020 Mar 31;11:226. doi: 10.3389/fgene.2020.00226. eCollection 2020. Front Genet. 2020. PMID: 32296456 Free PMC article. Review.
References
- Ziemert N; Alanjary M; Weber T The evolution of genome mining in microbes - a review. Nat. Prod. Rep. 2016, 33 (8), 988. - PubMed
- Tietz JI; Mitchell DA Using genomics for natural product structure elucidation. Curr. Top. Med. Chem. 2016, 16 (15), 1645. - PubMed
- Arnison PG; Bibb MJ; Bierbaum G; Bowers AA; Bugni TS; Bulaj G; Camarero JA; Campopiano DJ; Challis GL; Clardy J; Cotter PD; Craik DJ; Dawson M; Dittmann E; Donadio S; Dorrestein PC; Entian KD; Fischbach MA; Garavelli JS; Göransson U; Gruber CW; Haft DH; Hemscheidt TK; Hertweck C; Hill C; Horswill AR; Jaspars M; Kelly WL; Klinman JP; Kuipers OP; Link AJ; Liu W; Marahiel MA; Mitchell DA; Moll GN; Moore BS; Müller R; Nair SK; Nes IF; Norris GE; Olivera BM; Onaka H; Patchett ML; Piel J; Reaney MJ; Rebuffat S; Ross RP; Sahl HG; Schmidt EW; Selsted ME; Severinov K; Shen B; Sivonen K; Smith L; Stein T; Süssmuth RD; Tagg JR; Tang GL; Truman AW; Vederas JC; Walsh CT; Walton JD; Wenzel SC; Willey JM; van der Donk WA Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30 (1), 108. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources