Alzheimer's Disease Microbiome Is Associated with Dysregulation of the Anti-Inflammatory P-Glycoprotein Pathway - PubMed (original) (raw)
Alzheimer's Disease Microbiome Is Associated with Dysregulation of the Anti-Inflammatory P-Glycoprotein Pathway
John P Haran et al. mBio. 2019.
Abstract
The microbiota-gut-brain axis is a bidirectional communication system that is poorly understood. Alzheimer's disease (AD), the most common cause of dementia, has long been associated with bacterial infections and inflammation-causing immunosenescence. Recent studies examining the intestinal microbiota of AD patients revealed that their microbiome differs from that of subjects without dementia. In this work, we prospectively enrolled 108 nursing home elders and followed each for up to 5 months, collecting longitudinal stool samples from which we performed metagenomic sequencing and in vitro T84 intestinal epithelial cell functional assays for P-glycoprotein (P-gp) expression, a critical mediator of intestinal homeostasis. Our analysis identified clinical parameters as well as numerous microbial taxa and functional genes that act as predictors of AD dementia in comparison to elders without dementia or with other dementia types. We further demonstrate that stool samples from elders with AD can induce lower P-gp expression levels in vitro those samples from elders without dementia or with other dementia types. We also paired functional studies with machine learning approaches to identify bacterial species differentiating the microbiome of AD elders from that of elders without dementia, which in turn are accurate predictors of the loss of dysregulation of the P-gp pathway. We observed that the microbiome of AD elders shows a lower proportion and prevalence of bacteria with the potential to synthesize butyrate, as well as higher abundances of taxa that are known to cause proinflammatory states. Therefore, a potential nexus between the intestinal microbiome and AD is the modulation of intestinal homeostasis by increases in inflammatory, and decreases in anti-inflammatory, microbial metabolism.IMPORTANCE Studies of the intestinal microbiome and AD have demonstrated associations with microbiome composition at the genus level among matched cohorts. We move this body of literature forward by more deeply investigating microbiome composition via metagenomics and by comparing AD patients against those without dementia and with other dementia types. We also exploit machine learning approaches that combine both metagenomic and clinical data. Finally, our functional studies using stool samples from elders demonstrate how the c microbiome of AD elders can affect intestinal health via dysregulation of the P-glycoprotein pathway. P-glycoprotein dysregulation contributes directly to inflammatory disorders of the intestine. Since AD has been long thought to be linked to chronic bacterial infections as a possible etiology, our findings therefore fill a gap in knowledge in the field of AD research by identifying a nexus between the microbiome, loss of intestinal homeostasis, and inflammation that may underlie this neurodegenerative disorder.
Keywords: Alzheimer’s Disease; dementia; elderly; gut-brain axis; intestinal homeostasis; intestinal microbiome.
Copyright © 2019 Haran et al.
Figures
FIG 1
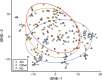
Microbiome diversity differs between Alzheimer’s disease elders and those with no dementia or other types of dementia. Stool samples from elders were sequenced via shotgun metagenomics. Samples were profiled for microbial species relative abundances by mapping reads to a NCBI bacterial genomes k-mer database with Kraken and by reconstructing the resulting relative abundance profile at the species levels with Bracken. Beta diversity was explored using Jaccard distances by _t_-distributed stochastic neighbor embedding (tSNE) for a measure of community species dissimilarity among samples collected from individuals without dementia (blue triangles), with Alzheimer’s disease (red circles), and with other dementia types (yellow squares). Each group is displayed with ellipses with a 95% confidence interval.
FIG 2
After adjusting for relative clinical covariates, microbiome composition differs at the genus level among elders with Alzheimer’s disease, no dementia, and other dementia types. We performed generalized mixed-effect modeling regression to predict genus-level proportions as a function of age, malnutrition, frailty, medications, and dementia state (no/other/AD). Patient ID was used as random effect to account for the repeated nature of our samples. Genera with greater than 0.1% mean relative abundance were significantly associated with Alzheimer’s disease (AD) or other dementia types (Other) in comparison to elders without dementia (NO). Only genera with a P value of <0.05 are presented, with relative abundance on the y axis.
FIG 3
Microbiome species composition, combined with frailty and malnutrition, accurately classify individuals as having Alzheimer’s disease versus no dementia. Random forest classification was performed according to Alzheimer’s disease versus no dementia (AD versus NO) by selecting at random one sample per individual (30 trials per run), from 100 starting random seeds, and building 3,000 decision trees per trial. (A) Out-of-bag (OOB) prediction error as a function of the number of decision trees run for elders without dementia (NO, blue) or with Alzheimer’s disease (AD, red). (B) Ranking of forest predictors based on average variable importance (e.g., mean decreased accuracy) across the 30 × 100 random trials. The top 30 important features discriminating the AD and NO dementia are reported. (C) Relative abundances for each species selected or scoring number for frailty and malnutrition clinical variables are reported with mean (thick bar) and up to minimum and maximum values for elders without dementia (NO, blue) or with Alzheimer’s disease (AD, red). In panel B, species are ordered based on importance.
FIG 4
Microbiome species composition accurately classify individuals as having Alzheimer’s disease versus other dementia types. Random forest classification was also performed according to Alzheimer’s disease versus no other dementia diagnosis (AD versus other) by selecting at random one sample per individual (30 trials per run), from 100 starting random seeds, and building 3,000 decision trees per trial. (A) Out-of-bag (OOB) prediction error as a function of the number of decision trees run for elders without dementia (other, yellow) or with Alzheimer’s disease (AD, red). (B) Ranking of forest predictors based on average variable importance (e.g., mean decreased accuracy) across the 30 × 100 random trials. The top 30 important features discriminating AD and other dementia are reported. (C) Relative abundances for each species selected by the model are reported with mean (thick bar) and up to minimum and maximum values (dots) for elders with other dementia types (other, yellow) or with Alzheimer’s disease (AD, red). In panel B, species are ordered based on importance.
FIG 5
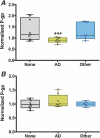
Intestinal microbiome of elders with Alzheimer’s disease induce significantly lower P-glycoprotein expression from intestinal epithelia cells, reflecting a higher level of inflammatory potential at the epithelial cell surface. (A and B) Supernatants were collected from stool samples from 9 randomly selected elders from each dementia classification group and incubated on T84 epithelial cells for 12 h prior to relative quantitation of P-gp by flow cytometry (A) or MRP2 by Western blotting (B). (A) P-glycoprotein expression was induced significantly less in samples from elders with Alzheimer’s disease (AD, yellow) than from those without dementia (none, gray) or with other dementia types (other, blue). Normalized P-glycoprotein (P-gp) expression is expressed on the y axis. (B) MRP2 did not show a significant difference in expression levels. ***, P < 0.05.
FIG 6
Dysbiotic microbiome species from elders with Alzheimer’s disease can accurately predict in vitro P-glycoprotein expression levels. Random forest regression analysis was performed to predict P-glycoprotein expression as a function of the bacterial relative abundances obtained from whole-genome sequencing of elders with Alzheimer’s disease and without any dementia. (A) A side-by-side comparison of the two models’ mean squared error (MSE) as a function of the number of generated trees demonstrates no difference between the MSE of the model obtained by training using only the top 30 features discriminating AD and no dementia (see Fig. 3, green line) and a model obtained by using information of all the available species (purple lines). The regression was performed in both cases starting from 500 different initial random seeds. (B) Variables with positive importance (and hence significantly contributing to the regression) resulted to be only a limited subset of taxa that differentiate Alzheimer’s disease from no-dementia individuals. (C) Species selected by the regression model are predicted to either induce or repress P-glycoprotein expression levels. Observations are colored according to dementia status (AD, red; no, blue).
Similar articles
- The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer's Disease-a Critical Review.
Sochocka M, Donskow-Łysoniewska K, Diniz BS, Kurpas D, Brzozowska E, Leszek J. Sochocka M, et al. Mol Neurobiol. 2019 Mar;56(3):1841-1851. doi: 10.1007/s12035-018-1188-4. Epub 2018 Jun 23. Mol Neurobiol. 2019. PMID: 29936690 Free PMC article. Review. - Altered microbiomes distinguish Alzheimer's disease from amnestic mild cognitive impairment and health in a Chinese cohort.
Liu P, Wu L, Peng G, Han Y, Tang R, Ge J, Zhang L, Jia L, Yue S, Zhou K, Li L, Luo B, Wang B. Liu P, et al. Brain Behav Immun. 2019 Aug;80:633-643. doi: 10.1016/j.bbi.2019.05.008. Epub 2019 May 4. Brain Behav Immun. 2019. PMID: 31063846 - Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer's disease.
Kesika P, Suganthy N, Sivamaruthi BS, Chaiyasut C. Kesika P, et al. Life Sci. 2021 Jan 1;264:118627. doi: 10.1016/j.lfs.2020.118627. Epub 2020 Oct 22. Life Sci. 2021. PMID: 33169684 Review. - Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis.
Song H, Yoo Y, Hwang J, Na YC, Kim HS. Song H, et al. J Allergy Clin Immunol. 2016 Mar;137(3):852-60. doi: 10.1016/j.jaci.2015.08.021. Epub 2015 Oct 1. J Allergy Clin Immunol. 2016. PMID: 26431583 - Disturbed microbial ecology in Alzheimer's disease: evidence from the gut microbiota and fecal metabolome.
Xi J, Ding D, Zhu H, Wang R, Su F, Wu W, Xiao Z, Liang X, Zhao Q, Hong Z, Fu H, Xiao Q. Xi J, et al. BMC Microbiol. 2021 Aug 12;21(1):226. doi: 10.1186/s12866-021-02286-z. BMC Microbiol. 2021. PMID: 34384375 Free PMC article.
Cited by
- Gut Microbiota Alterations in Alzheimer's Disease: Relation with Cognitive Impairment and Mediterranean Lifestyle.
Mateo D, Carrión N, Cabrera C, Heredia L, Marquès M, Forcadell-Ferreres E, Pino M, Zaragoza J, Moral A, Cavallé L, González-de-Echávarri JM, Vicens P, Domingo JL, Torrente M. Mateo D, et al. Microorganisms. 2024 Oct 10;12(10):2046. doi: 10.3390/microorganisms12102046. Microorganisms. 2024. PMID: 39458354 Free PMC article. - Gut Microbiome in Alzheimer's Disease: from Mice to Humans.
Liang C, Pereira R, Zhang Y, Rojas OL. Liang C, et al. Curr Neuropharmacol. 2024;22(14):2314-2329. doi: 10.2174/1570159X22666240308090741. Curr Neuropharmacol. 2024. PMID: 39403057 Free PMC article. Review. - Identification of proteotoxic and proteoprotective bacteria that non-specifically affect proteins associated with neurodegenerative diseases.
Walker AC, Bhargava R, Bucher MJ, Argote YM, Brust AS, Czyż DM. Walker AC, et al. iScience. 2024 Aug 27;27(9):110828. doi: 10.1016/j.isci.2024.110828. eCollection 2024 Sep 20. iScience. 2024. PMID: 39310761 Free PMC article. - Gut Microbiome Compositional and Functional Features Associate with Alzheimer's Disease Pathology.
Kang JW, Khatib LA, Heston MB, Dilmore AH, Labus JS, Deming Y, Schimmel L, Blach C, McDonald D, Gonzalez A, Bryant M, Sanders K, Schwartz A, Ulland TK, Johnson SC, Asthana S, Carlsson CM, Chin NA, Blennow K, Zetterberg H, Rey FE; Alzheimer Gut Microbiome Project Consortium; Kaddurah-Daouk R, Knight R, Bendlin BB. Kang JW, et al. medRxiv [Preprint]. 2024 Sep 5:2024.09.04.24313004. doi: 10.1101/2024.09.04.24313004. medRxiv. 2024. PMID: 39281749 Free PMC article. Preprint. - Causal Association between Circulating Metabolites and Dementia: A Mendelian Randomization Study.
Li HM, Qiu CS, Du LY, Tang XL, Liao DQ, Xiong ZY, Lai SM, Huang HX, Kuang L, Zhang BY, Li ZH. Li HM, et al. Nutrients. 2024 Aug 28;16(17):2879. doi: 10.3390/nu16172879. Nutrients. 2024. PMID: 39275195 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK056754/DK/NIDDK NIH HHS/United States
- R03 AG056356/AG/NIA NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- T32 AI007349/AI/NIAID NIH HHS/United States
- K23 AG057790/AG/NIA NIH HHS/United States
- R15 AI112985/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous