Effect of Disease-Associated Germline Mutations on Structure Function Relationship of DNA Methyltransferases - PubMed (original) (raw)
Review
Effect of Disease-Associated Germline Mutations on Structure Function Relationship of DNA Methyltransferases
Allison B Norvil et al. Genes (Basel). 2019.
Abstract
Despite a large body of evidence supporting the role of aberrant DNA methylation in etiology of several human diseases, the fundamental mechanisms that regulate the activity of mammalian DNA methyltransferases (DNMTs) are not fully understood. Recent advances in whole genome association studies have helped identify mutations and genetic alterations of DNMTs in various diseases that have a potential to affect the biological function and activity of these enzymes. Several of these mutations are germline-transmitted and associated with a number of hereditary disorders, which are potentially caused by aberrant DNA methylation patterns in the regulatory compartments of the genome. These hereditary disorders usually cause neurological dysfunction, growth defects, and inherited cancers. Biochemical and biological characterization of DNMT variants can reveal the molecular mechanism of these enzymes and give insights on their specific functions. In this review, we introduce roles and regulation of DNA methylation and DNMTs. We discuss DNMT mutations that are associated with rare diseases, the characterized effects of these mutations on enzyme activity and provide insights on their potential effects based on the known crystal structure of these proteins.
Keywords: ADCA-DN; DNA methylation; DNMT1; DNMT3A; HSAN1E; PCC/PGL; TBRS; dwarfism; rare diseases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
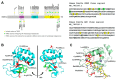
HSAN1E (hereditary sensory neuropathy with dementia and hearing loss) and ADCA-DN (autosomal dominant cerebellar ataxia, deafness and narcolepsy) mutations in DNMT1 (A) Left Schematic representation of the hDNMT1 gene. HSAN1E mutations are listed above the gene in light blue, while ADCA-DN mutations are listed below the gene in red. Right Nucleotide sequence of the RFTS (replication foci-targeting sequence) domain, with the mutations highlighted in color corresponding to the schematic. (B) Crystal structure of hDNMT1 (351–1600) from the Protein Data Bank (PDB: 4WXX). The cartoon structure of the RFTS domain is green, the CXXC domain is purple, and the MTase (methyltransferase) domain is yellow. All disease mutations are located in the RFTS domain, and are shown as stick structures in red. The positions of HSAN1E and ADCA-DN mutations are shown in the left and right DNMT1 structure respectively. (C) Overlay of the hDNMT1 RFTS domain bound (light blue) and unbound (green) to two molecules of ubiquitin (dark blue) from the PDB: 4WXX and 5YDR. When ubiquitin is bound, the RFTS domain bends about 30° at Met502. The HSAN1E mutation Lys505del and Met502 are shown in red in RFTS domain bound to ubiquitin and in orange in the unbound RFTS domain. (D) Model showing the effect of mutations in the RFTS domain on the catalytic mechanism of DNMT1. DNMT1 is auto-inhibited by the interaction of its RFTS domain with the target recognition domain (TRD) in MTase domain that prevents DNA binding. When RFTS interacts with ubiquitin, auto-inhibition is released allowing TRD to interact with the hemi-methylated DNA. However, mutations in the RFTS that alter its binding to ubiquitin will prevent enzyme activation, while mutations that alter its binding to the TRD will leave the enzyme in a hyperactive state.
Figure 2
TBRS (Tatton-Brown-Rahman syndrome) mutations in the PWWP and ADD domain of DNMT3A (A) Left Schematic representation of the hDNMT3A gene, with mutations listed in different domains. Variants unique to TBRS are highlighted in yellow, TBRS variants overlapping with hematologic malignancies are highlighted in green, and TBRS variants at the codon that are altered to different amino acids in hematologic malignancies are highlighted in grey. In the schematic, (*) is used to indicate a stop codon replacement, Ffs indicates a frame-shift mutation, and CS indicates the catalytic site. Right Nucleotide sequence of the PWWP (Pro-Trp-Trp-Pro) and ADD (ATRX-DNMT3-DNMT3L) domain, with the mutations highlighted in the color corresponding to the schematic. (B) Two orientations of the DNMT3A PWWP domain (PDB: 3LLR). The positions of TBRS mutations are shown as stick structures in red, whereas residues part of the aromatic cage that bind to H3K36me2/3 shown as stick structures in green. (C) Crystal Structure of the DNMT3A ADD domain (PDB: 4U7T), in green bound to an unmodified H3 peptide shown as a stick structure in yellow. Grey spheres represent bound zinc. The positions of TBRS mutations are shown as a stick structure in red.
Figure 3
TBRS mutations in the catalytic domain of DNMT3A (A) Nucleotide sequence of the hDNMT3A MTase domain. As seen in Figure 2A, variants unique to TBRS are highlighted in yellow, TBRS variants overlapping with hematologic malignancies are highlighted in green, and TBRS variants at the codon that are altered to different amino acids in hematologic malignancies are highlighted in grey. (B) Crystal structure of DNMT3A bound to DNA, zoomed in to show only one of the two monomers in the tetrameric structure (PDB: 5YX2). The MTase domain is shown in a pink cartoon structure and the positions of TBRS mutations are shown as stick structures in red. The second monomer of the MTase is shown in grey. (C–G) Magnified view of the motifs I-III (C), motif IV (D–E), motif VIII (F), the TRD (G), and motif X (H) shown as stick structures in blue and the positions of TBRS mutations shown as stick structures in red. The black dotted lines represent interactions with nearby residues or with the DNA. (H) The interface between DNMT3A, pink cartoon, and DNMT3L, orange cartoon. The positions of TBRS mutations shown as stick structures in red. The black dotted lines represent interactions with nearby residues of DNMT3L shown as stick structures in orange.
Figure 4
PCC/PGL (Pheochromocytoma/paraganglioma) and MD (microcephalic dwarfism) mutations in PWWP domain of DNMT3A (A,C) Top Schematic representation of the hDNMT3A gene, showing PCC/PGL and MD mutations in the PWWP domain. Below Nucleotide sequence of the PWWP domain, with the mutations highlighted in pink and blue respectively. (B,D) Crystal structure of DNMT3A PWWP domain shown in blue (PDB: 3LLR). The positions of PCC/PGL mutation (B) and MD mutations (D) shown as stick structures in red. The aromatic cage residues are shown in green.
Similar articles
- Aberrant signature methylome by DNMT1 hot spot mutation in hereditary sensory and autonomic neuropathy 1E.
Sun Z, Wu Y, Ordog T, Baheti S, Nie J, Duan X, Hojo K, Kocher JP, Dyck PJ, Klein CJ. Sun Z, et al. Epigenetics. 2014 Aug;9(8):1184-93. doi: 10.4161/epi.29676. Epub 2014 Jul 7. Epigenetics. 2014. PMID: 25033457 Free PMC article. - Autosomal dominant cerebellar ataxia, deafness, and narcolepsy (ADCA-DN) associated with progressive cognitive and behavioral deterioration.
Walker LA, Bourque P, Smith AM, Warman Chardon J. Walker LA, et al. Neuropsychology. 2017 Mar;31(3):292-303. doi: 10.1037/neu0000322. Epub 2016 Nov 21. Neuropsychology. 2017. PMID: 27869457 - [DNA methyltransferases: classification, functions and research progress].
Wang ZG, Wu JX. Wang ZG, et al. Yi Chuan. 2009 Sep;31(9):903-12. doi: 10.3724/sp.j.1005.2009.00903. Yi Chuan. 2009. PMID: 19819843 Review. Chinese. - DNA methyltransferases in hematological malignancies.
Hoang NM, Rui L. Hoang NM, et al. J Genet Genomics. 2020 Jul 20;47(7):361-372. doi: 10.1016/j.jgg.2020.04.006. Epub 2020 Jul 24. J Genet Genomics. 2020. PMID: 32994141 Free PMC article. Review.
Cited by
- F-box protein CFK1 interacts with and degrades de novo DNA methyltransferase in Arabidopsis.
Chen J, Liu J, Jiang J, Qian S, Song J, Kabara R, Delo I, Serino G, Liu F, Hua Z, Zhong X. Chen J, et al. New Phytol. 2021 Mar;229(6):3303-3317. doi: 10.1111/nph.17103. Epub 2020 Dec 15. New Phytol. 2021. PMID: 33216996 Free PMC article. - Functional Analysis of DNMT3A DNA Methyltransferase Mutations Reported in Patients with Acute Myeloid Leukemia.
Khrabrova DA, Loiko AG, Tolkacheva AA, Cherepanova NA, Zvereva MI, Kirsanova OV, Gromova ES. Khrabrova DA, et al. Biomolecules. 2019 Dec 18;10(1):8. doi: 10.3390/biom10010008. Biomolecules. 2019. PMID: 31861499 Free PMC article. - Both intra and inter-domain interactions define the intrinsic dynamics and allosteric mechanism in DNMT1s.
Liang Z, Zhu Y, Long J, Ye F, Hu G. Liang Z, et al. Comput Struct Biotechnol J. 2020 Mar 23;18:749-764. doi: 10.1016/j.csbj.2020.03.016. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32280430 Free PMC article. - The acute myeloid leukemia variant DNMT3A Arg882His is a DNMT3B-like enzyme.
Norvil AB, AlAbdi L, Liu B, Tu YH, Forstoffer NE, Michie AR, Chen T, Gowher H. Norvil AB, et al. Nucleic Acids Res. 2020 Apr 17;48(7):3761-3775. doi: 10.1093/nar/gkaa139. Nucleic Acids Res. 2020. PMID: 32123902 Free PMC article. - Disease-Associated Mutation A554V Disrupts Normal Autoinhibition of DNMT1.
Switzer RL, Hartman ZJ, Hewett GR, Carroll CF. Switzer RL, et al. DNA (Basel). 2023 Sep;3(3):119-133. doi: 10.3390/dna3030010. Epub 2023 Jul 13. DNA (Basel). 2023. PMID: 37663147 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources