A newly isolated human intestinal bacterium strain capable of deglycosylating flavone C-glycosides and its functional properties - PubMed (original) (raw)
A newly isolated human intestinal bacterium strain capable of deglycosylating flavone C-glycosides and its functional properties
Shiqi Zheng et al. Microb Cell Fact. 2019.
Abstract
Background: Flavone C-glycosides are difficult to be deglycosylated using traditional chemical methods due to their solid carbon-carbon bond between sugar moieties and aglycones; however, some bacteria may easily cleave this bond because they generate various specific enzymes.
Results: A bacterial strain, named W12-1, capable of deglycosylating orientin, vitexin, and isovitexin to their aglycones, was isolated from human intestinal bacteria in this study and identified as Enterococcus faecalis based on morphological examination, physiological and biochemical identification, and 16S rDNA sequencing. The strain was shown to preferentially deglycosylate the flavone C-glycosides on condition that the culture medium was short of carbon nutrition sources such as glucose and starch, and its deglycosylation efficiency was negatively correlated with the content of the latter two substances.
Conclusion: This study provided a new bacterial resource for the cleavage of C-glycosidic bond of flavone C-glycosides and reported the carbon nutrition sources reduction induced deglycosylation for the first time.
Keywords: Deglycosylation; Enterococcus faecalis; Flavone C-glycosides; Reduced carbon nutrition source induction.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Fig. 1
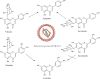
Structures and transformation of the compounds of interest
Fig. 2
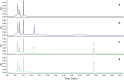
HPLC chromatograms related to deglycosylation of orientin. a blank group; b experimental group at 0 h; c reference compounds of orientin and luteolin in GAM; d experimental group at 24 h. 1: orientin; 2: luteolin
Fig. 3
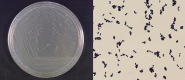
Morphological characteristics of strain W12-1. Left: the colony; Right: the cells
Fig. 4
Neighbor-joining tree based on 16S rDNA gene sequences
Fig. 5
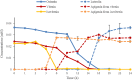
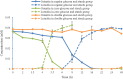
Time course of deglycosylation of orientin, vitexin and isovitexin (n = 3, mean ± SD)
Fig. 6
Time course of deglycosylation of orientin in the media with different contents of carbon nutrition (n = 3, mean ± SD)
Fig. 7
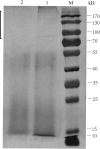
Comparative SDS-PAGE electrophoretogram of strain W 12-1 culture in the media with double content of carbon sources and without carbon sources. M: protein ladder; 1: the culture in the medium without carbon sources; 2: the culture in the medium with double content level of carbon sources
Similar articles
- Isolation and identification of Enterococcus gallinarum P581a, a strain of intestinal bacteria deglycosylating flavone C-glycosides.
Tao Z, Zheng S, Liu S, Wang J, Geng D, Wang R. Tao Z, et al. J Gen Appl Microbiol. 2022 Nov 10;68(3):125-133. doi: 10.2323/jgam.2021.10.002. Epub 2022 May 31. J Gen Appl Microbiol. 2022. PMID: 35650022 - A newly isolated human intestinal strain deglycosylating flavonoid C-glycosides.
Wang S, Liu S, Wang J, Tao J, Wu M, Ma W, Wang R. Wang S, et al. Arch Microbiol. 2022 May 10;204(6):310. doi: 10.1007/s00203-022-02881-2. Arch Microbiol. 2022. PMID: 35536516 - Deglycosylation of isoflavone C-glycosides by newly isolated human intestinal bacteria.
Kim M, Lee J, Han J. Kim M, et al. J Sci Food Agric. 2015 Jul;95(9):1925-31. doi: 10.1002/jsfa.6900. Epub 2014 Sep 30. J Sci Food Agric. 2015. PMID: 25199800 - Discovery and mechanism of intestinal bacteria in enzymatic cleavage of C-C glycosidic bonds.
Wei B, Wang YK, Qiu WH, Wang SJ, Wu YH, Xu XW, Wang H. Wei B, et al. Appl Microbiol Biotechnol. 2020 Mar;104(5):1883-1890. doi: 10.1007/s00253-019-10333-z. Epub 2020 Jan 13. Appl Microbiol Biotechnol. 2020. PMID: 31932892 Review. - Microbial biotransformation of bioactive flavonoids.
Cao H, Chen X, Jassbi AR, Xiao J. Cao H, et al. Biotechnol Adv. 2015 Jan-Feb;33(1):214-223. doi: 10.1016/j.biotechadv.2014.10.012. Epub 2014 Nov 4. Biotechnol Adv. 2015. PMID: 25447420 Review.
Cited by
- An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nasseri SA, Lazarski AC, Lemmer IL, Zhang CY, Brencher E, Chen HM, Sim L, Panwar D, Betschart L, Worrall LJ, Brumer H, Strynadka NCJ, Withers SG. Nasseri SA, et al. Nature. 2024 Jul;631(8019):199-206. doi: 10.1038/s41586-024-07574-y. Epub 2024 Jun 19. Nature. 2024. PMID: 38898276 - Genomics and physiology of Catenibacillus, human gut bacteria capable of polyphenol _C_-deglycosylation and flavonoid degradation.
Goris T, Braune A. Goris T, et al. Microb Genom. 2024 May;10(5):001245. doi: 10.1099/mgen.0.001245. Microb Genom. 2024. PMID: 38785231 Free PMC article. - Anti-Proliferative Potential of Cynaroside and Orientin-In Silico (DYRK2) and In Vitro (U87 and Caco-2) Studies.
Pirvu LC, Pintilie L, Albulescu A, Stefaniu A, Neagu G. Pirvu LC, et al. Int J Mol Sci. 2023 Nov 21;24(23):16555. doi: 10.3390/ijms242316555. Int J Mol Sci. 2023. PMID: 38068880 Free PMC article. - Enzymatic β-elimination in natural product O- and C-glycoside deglycosylation.
Bitter J, Pfeiffer M, Borg AJE, Kuhlmann K, Pavkov-Keller T, Sánchez-Murcia PA, Nidetzky B. Bitter J, et al. Nat Commun. 2023 Nov 6;14(1):7123. doi: 10.1038/s41467-023-42750-0. Nat Commun. 2023. PMID: 37932298 Free PMC article. - _C_-Glycoside-Metabolizing Human Gut Bacterium, Dorea sp. MRG-IFC3.
Mi HTN, Chaiyasarn S, Kim H, Han J. Mi HTN, et al. J Microbiol Biotechnol. 2023 Dec 28;33(12):1606-1614. doi: 10.4014/jmb.2308.08021. Epub 2023 Sep 21. J Microbiol Biotechnol. 2023. PMID: 37789701 Free PMC article.
References
- Leska M, Hiller K. Flavone C-glycosides: a review. Pharmazie. 1988;43:305–312.
- Wu XA, Zhao YM. Advances in the research of natural flavonoid C-glycosides and their activities. Pharm J Chin PLA. 2005;21:135–138.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical