Microbial Selection and Survival in Subseafloor Sediment - PubMed (original) (raw)
Microbial Selection and Survival in Subseafloor Sediment
John B Kirkpatrick et al. Front Microbiol. 2019.
Abstract
Many studies have examined relationships of microorganisms to geochemical zones in subseafloor sediment. However, responses to selective pressure and patterns of community succession with sediment depth have rarely been examined. Here we use 16S rDNA sequencing to examine the succession of microbial communities at sites in the Indian Ocean and the Bering Sea. The sediment ranges in depth from 0.16 to 332 m below seafloor and in age from 660 to 1,300,000 years. The majority of subseafloor taxonomic diversity is present in the shallowest depth sampled. The best predictor of sequence presence or absence in the oldest sediment is relative abundance in the near-seafloor sediment. This relationship suggests that perseverance of specific taxa into deep, old sediment is primarily controlled by the taxonomic abundance that existed when the sediment was near the seafloor. The operational taxonomic units that dominate at depth comprise a subset of the local seafloor community at each site, rather than a grown-in group of geographically widespread subseafloor specialists. At both sites, most taxa classified as abundant decrease in relative frequency with increasing sediment depth and age. Comparison of community composition to cell counts at the Bering Sea site indicates that the rise of the few dominant taxa in the deep subseafloor community does not require net replication, but might simply result from lower mortality relative to competing taxa on the long timescale of community burial.
Keywords: 16S rDNA; NGHP-14; U1343; deep biosphere; marine sediment archaea; marine sediment bacteria; microbial selection.
Figures
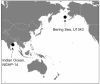
FIGURE 1
Sampling locations (Bering Sea IODP Site U1343 and Bay of Bengal NGHP Site 14).
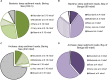
FIGURE 2
Persistence of abundant, moderate, rare, and total OTUs at depth (meters below seafloor, mbsf), plotted as a percentage of the OTU count for each category in the shallowest sample (“near seafloor”). All categories total 100% for the sample nearest the seafloor, where the number of OTUs in each category is greatest. Circles represent subgroups of bacterial OTUs and squares represent archaea. (A) Bering Sea (Site U1343) Bacteria, (B) Bay of Bengal (NGHP-14) Bacteria, (C) Bering Sea (U1343) Archaea, and (D) Bay of Bengal (NGHP-14) Archaea. For (A,B), the shallowest sample is at 0.16 mbsf. For (C,D), the shallowest sample is at 4.0 mbsf.
FIGURE 3
Composition of the deepest sediment communities sampled, as identified by OTU abundance or absence in the shallowest sample from each site. (A) Bacteria from 332 mbsf at Bering Sea Site U1343, (B) Bacteria from 69 mbsf at Bay of Bengal Site NGHP-14, (C) Archaea from 332 mbsf at U1343, (D) Archaea at 69 mbsf from NGHP-14.
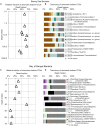
FIGURE 4
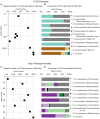
Relative read-based abundance in each sample of bacterial OTUs that are abundant in the shallowest sample at each site. Abundant OTUs are those that comprise more than 1% of the total reads from the shallowest sample. (A) For Bering Sea Site U1343, the fraction of total reads represented by all of these OTUs at each sampled depth. (B) The fraction of reads represented by each of these OTUs in depth order, with taxonomy. Taxonomic assignments are listed as phylum, and then lowest identifiable level in parentheses (default is genus). “Ca.” is Candidatus. For complete taxonomy see Supplemental Tables 1, 2. (C) For Indian Ocean Site NGHP-14, the fraction of reads represented by all of these OTUs at each sampled depth. (D) The fraction of reads represented by each of these OTUs in depth order, including taxonomy (format same as B). Color palette for this figure, Figure 5–9 informed by ColorBrewer 2.0 (Harrower and Brewer, 2003).
FIGURE 5
Relative read-based abundance in each sample of archaeal OTUs that are abundant in the shallowest sample at each site. Abundant OTUs are those that comprise more than 1% of the total reads from the shallowest sample. (A) For Bering Sea Site U1343, the fraction of total reads represented by all of these OTUs at each sampled depth. (B) The fraction of reads represented by each of these OTUs in depth order, with taxonomy. Taxonomic assignments are listed as phylum, and then lowest identifiable level in parentheses (default is genus). “Ca.” is Candidatus, “un. ge.” is uncultured genus. For complete taxonomy see Supplementary Tables 1, 2. (C) For Indian Ocean Site NGHP-14, the fraction of reads represented by all of these OTUs at each sampled depth. (D) The fraction of reads represented by each of these OTUs in depth order, including taxonomy (format same as B).
FIGURE 6
Relative read-based abundance in each horizon of bacterial taxa that dominate the deepest sample. Deep dominant OTUs are OTUs represented by >1% of the sequences returned from the deepest sample. (A) The fraction of bacterial reads comprised by all of the deep dominant OTUs at each depth for Bering Sea site U1343. (B) The fraction of reads represented by each of these OTUs in depth order, with taxonomy. Taxonomic assignments are listed as phylum, and then lowest identifiable level in parentheses (default is genus). “Ca.” is Candidatus. For complete taxonomy see Supplementary Tables 1, 2. (C) The fraction of bacterial reads comprised by all of the deep dominant OTUs at each depth for Bay of Bengal site NGHP-14. (D) The fraction of reads represented by each of these OTUs in depth order, including taxonomy.
FIGURE 7
Relative read-based abundance in each horizon of archaeal taxa that dominate the deepest sample. Deep dominant OTUs are OTUs represented by >1% of the sequences returned from the deepest sample. (A) The fraction of archaeal reads comprised by all of the deep dominant OTUs at each depth for Bering Sea site U1343. (B) Archaeal taxonomy and relative abundance in depth order. Taxonomic assignments are listed as phylum, and then lowest identifiable level in parentheses (default is genus). “Ca.” is Candidatus, “un. ge.” is uncultured genus. For complete taxonomy see Supplementary Tables 1, 2. (C) The fraction of archaeal reads comprised by all of the deep dominant OTUs at each depth for Bay of Bengal site NGHP-14. (D) Archaeal taxonomy and relative abundance in depth order. Format same as (B).
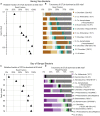
FIGURE 8
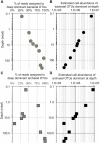
The relative fraction of reads at each depth assigned to individual bacterial OTUs (97% similarity cutoff) that are abundant in the shallowest sample at (A) Bering Sea Site U1343 and (B) Bay of Bengal Site NGHP-14. The dashed line represents 100% of surface reads (i.e., no change in relative abundance with depth). Values to the right of the dashed line indicate higher frequency of the sequences assigned to each OTU (relative to the shallowest sample), whereas values to the left indicate lower frequency relative to the shallowest sample. Rank order is the same as Figure 4. For reference, for (A) rank order 5 is Ca. “Aerophobetes,” 9 is Chloroflexi (Anaerolinaceae), and 13 is Chloroflexi (Dehalococcoidia; “DscP2”). For (B) rank order 5 is Chloroflexi (Dehalococcoidia; “DscP2”) and 9 is Ca. “Atribacteria” (JS1). Axes are logarithmic. Values to the left of the y-axis represent “zero” (not detected).
FIGURE 9
The relative fraction of reads assigned to individual archaeal OTUs (97% similarity cutoff) that are abundant in the shallowest sample at (A) Bering Sea Site U1343 and (B) Bay of Bengal Site NGHP-14. The dashed line represents 100% of surface reads (no change in relative abundance with depth). Values to the right of the dashed line indicate higher frequency of the sequences assigned to each OTU (relative to the shallowest sample), whereas values to the left indicate lower frequency relative to the shallowest sample. Rank order same as Figure 5. For reference, for (A) rank order 1 is “Bathyarchaeia,” and 4 and 6 also fall in this group. For (B) rank order 3 is in “Bathyarchaeia” and so is 4. Axes are logarithmic. Values to the left of the y-axis represent “zero” (not detected).
FIGURE 10
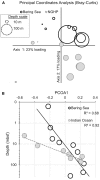
Principal coordinates analysis (PCoA) for bacteria showing the similarity of samples from the two different sites and different depths. Similarity was determined by abundance-weighted metrics (Bray–Curtis). (A) PCoA plot. The loading explained by the two axes is shown (23% for x, 17% for y). Symbol size represents sample depth. (B) Primary axis ordination (23% of variance) versus sample depth, with an exponential fit. Open symbols with solid outlines indicate Bering Sea samples. Gray symbols with dashed outlines indicate Bay of Bengal samples.
FIGURE 11
Comparison of the increase in OTU dominance to the decrease in cell abundance with increasing depth at Bering Sea Site U1343. (A) The fraction of reads contained in a bin of the 10 bacterial OTUs that are most abundant in the deepest samples at Bering Sea (Site U1343). (B) The calculated number of cells represented by these OTUs versus depth (logarithmic scale), determined from (A) and the trendline of U1343 cell counts with depth (cell counts from Kallmeyer et al., 2012). (C) The fraction of reads contained in a bin of the 10 archaeal OTUs that are most abundant in the deepest sample at Site U1343. (D) Similar to (B), the calculated number of cells represented by these OTUs versus depth (logarithmic scale).
Similar articles
- Influence of 16S rRNA Hypervariable Region on Estimates of Bacterial Diversity and Community Composition in Seawater and Marine Sediment.
Kerrigan Z, Kirkpatrick JB, D'Hondt S. Kerrigan Z, et al. Front Microbiol. 2019 Jul 16;10:1640. doi: 10.3389/fmicb.2019.01640. eCollection 2019. Front Microbiol. 2019. PMID: 31379788 Free PMC article. - Relationship of Bacterial Richness to Organic Degradation Rate and Sediment Age in Subseafloor Sediment.
Walsh EA, Kirkpatrick JB, Pockalny R, Sauvage J, Spivack AJ, Murray RW, Sogin ML, D'Hondt S. Walsh EA, et al. Appl Environ Microbiol. 2016 Jul 29;82(16):4994-9. doi: 10.1128/AEM.00809-16. Print 2016 Aug 15. Appl Environ Microbiol. 2016. PMID: 27287321 Free PMC article. - Bacterial diversity and community composition from seasurface to subseafloor.
Walsh EA, Kirkpatrick JB, Rutherford SD, Smith DC, Sogin M, D'Hondt S. Walsh EA, et al. ISME J. 2016 Apr;10(4):979-89. doi: 10.1038/ismej.2015.175. Epub 2015 Oct 2. ISME J. 2016. PMID: 26430855 Free PMC article. - Atribacteria Reproducing over Millions of Years in the Atlantic Abyssal Subseafloor.
Vuillemin A, Vargas S, Coskun ÖK, Pockalny R, Murray RW, Smith DC, D'Hondt S, Orsi WD. Vuillemin A, et al. mBio. 2020 Oct 6;11(5):e01937-20. doi: 10.1128/mBio.01937-20. mBio. 2020. PMID: 33024037 Free PMC article. - Prokaryotic biodiversity and activity in the deep subseafloor biosphere.
Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G. Fry JC, et al. FEMS Microbiol Ecol. 2008 Nov;66(2):181-96. doi: 10.1111/j.1574-6941.2008.00566.x. Epub 2008 Aug 20. FEMS Microbiol Ecol. 2008. PMID: 18752622 Review.
Cited by
- Microbial community structure in hadal sediments: high similarity along trench axes and strong changes along redox gradients.
Schauberger C, Glud RN, Hausmann B, Trouche B, Maignien L, Poulain J, Wincker P, Arnaud-Haond S, Wenzhöfer F, Thamdrup B. Schauberger C, et al. ISME J. 2021 Dec;15(12):3455-3467. doi: 10.1038/s41396-021-01021-w. Epub 2021 Jun 8. ISME J. 2021. PMID: 34103697 Free PMC article. - Influence of 16S rRNA Hypervariable Region on Estimates of Bacterial Diversity and Community Composition in Seawater and Marine Sediment.
Kerrigan Z, Kirkpatrick JB, D'Hondt S. Kerrigan Z, et al. Front Microbiol. 2019 Jul 16;10:1640. doi: 10.3389/fmicb.2019.01640. eCollection 2019. Front Microbiol. 2019. PMID: 31379788 Free PMC article. - Genomic insights into Penicillium chrysogenum adaptation to subseafloor sedimentary environments.
Liu X, Wang X, Zhou F, Xue Y, Liu C. Liu X, et al. BMC Genomics. 2024 Jan 2;25(1):4. doi: 10.1186/s12864-023-09921-1. BMC Genomics. 2024. PMID: 38166640 Free PMC article. - Subseafloor life and its biogeochemical impacts.
D'Hondt S, Pockalny R, Fulfer VM, Spivack AJ. D'Hondt S, et al. Nat Commun. 2019 Aug 6;10(1):3519. doi: 10.1038/s41467-019-11450-z. Nat Commun. 2019. PMID: 31388058 Free PMC article. Review. - The Guaymas Basin Subseafloor Sedimentary Archaeome Reflects Complex Environmental Histories.
Ramírez GA, McKay LJ, Fields MW, Buckley A, Mortera C, Hensen C, Ravelo AC, Teske AP. Ramírez GA, et al. iScience. 2020 Aug 15;23(9):101459. doi: 10.1016/j.isci.2020.101459. eCollection 2020 Sep 25. iScience. 2020. PMID: 32861995 Free PMC article.
References
- Bray J. R., Curtis J. T. (1957). An ordination of the upland forest communities of southern wisconsin. Ecol. Monogr. 27 325–349. 10.2307/1942268 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources