ProteinNet: a standardized data set for machine learning of protein structure - PubMed (original) (raw)
ProteinNet: a standardized data set for machine learning of protein structure
Mohammed AlQuraishi. BMC Bioinformatics. 2019.
Abstract
Background: Rapid progress in deep learning has spurred its application to bioinformatics problems including protein structure prediction and design. In classic machine learning problems like computer vision, progress has been driven by standardized data sets that facilitate fair assessment of new methods and lower the barrier to entry for non-domain experts. While data sets of protein sequence and structure exist, they lack certain components critical for machine learning, including high-quality multiple sequence alignments and insulated training/validation splits that account for deep but only weakly detectable homology across protein space.
Results: We created the ProteinNet series of data sets to provide a standardized mechanism for training and assessing data-driven models of protein sequence-structure relationships. ProteinNet integrates sequence, structure, and evolutionary information in programmatically accessible file formats tailored for machine learning frameworks. Multiple sequence alignments of all structurally characterized proteins were created using substantial high-performance computing resources. Standardized data splits were also generated to emulate the difficulty of past CASP (Critical Assessment of protein Structure Prediction) experiments by resetting protein sequence and structure space to the historical states that preceded six prior CASPs. Utilizing sensitive evolution-based distance metrics to segregate distantly related proteins, we have additionally created validation sets distinct from the official CASP sets that faithfully mimic their difficulty.
Conclusion: ProteinNet represents a comprehensive and accessible resource for training and assessing machine-learned models of protein structure.
Keywords: CASP; Co-evolution; Database; Deep learning; Machine learning; PSSM; Protein sequence; Protein structure; Protein structure prediction; Proteins.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Fig. 1
ProteinNet construction pipeline. For each ProteinNet, all proteins with PDB structures available prior to the start of its corresponding CASP (“All data”, top circle) are clustered using an MSA-based clustering technique (left inset) to yield large clusters where intra-cluster sequence identity is as low as 10%. One exemplar from each cluster is then selected (right inset) to yield the 10% seq. id. validation set. This process is iteratively repeated, by reclustering the data remaining outside of all initial clusters to yield validation sets of higher sequence identity (20–90%). Once the final validation set is extracted, all remaining data is used to form the training set. Based on this set (“100% thinning”), filtered training sets are created at lower sequence identity thresholds to provide coarser sampling of sequence space. Left inset: Each protein sequence is queried against a large sequence database (filtered to only include sequences publicly available prior to the beginning of the corresponding CASP) using JackHMMer to create an MSA that is subsequently filtered to 90% seq. id. HHblits is then used to perform an all-against-all sequence alignment of MSAs. Finally, alignment distances are fed to MMseqs2 to cluster their corresponding sequences. Right inset: The center-most protein of each cluster is chosen to ensure that the desired sequence identity constraints are satisfied, as proteins near cluster boundaries may be closer than the pre-specified radius of each cluster (pink vs. gray measuring tapes), while the distances between cluster centroids must satisfy the sequence identity constraints (blue measuring tape). The centroids are then used to form tight clusters of 95% seq. id. that are intersected with the original clusters to yield candidate exemplars ranked by multiple quality metrics (see main text). The top-ranked candidate is picked as the exemplar protein of each cluster
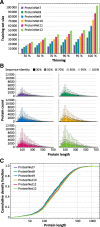
Fig. 2
Statistics of ProteinNet data sets. a Number of proteins in ProteinNet training sets for different thinnings (30–100% seq. id.) (b) Protein length distributions for ProteinNet training sets. c Cumulative density function of protein length distribution for 100% thinnings of ProteinNet training sets
Fig. 3
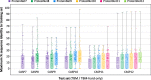
Alignment size as a function of ProteinNet subset. Box and whisker charts depict the distribution of number of sequences per MSA for ProteinNet training (30% thinning), validation, and test sets. Individual data points for training sets are not shown due to their large size
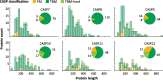
Fig. 4
Statistics of CASP data sets. Length distribution of proteins in CASP 7 through 12, broken down by difficulty class. Pie charts show the number of proteins per difficulty class
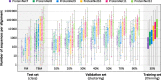
Fig. 5
Distributions of maximum % sequence identity to training sets. Box and whisker charts depict the distribution of maximum % sequence identity, with respect to the training set, of all entries in a given ProteinNet validation or test set. The FM test sets and 10% seq. id. validation sets show a median value of 0% seq. id. to the training set
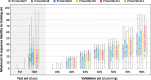
Fig. 6
Distributions of maximum % sequence identity of CASP entries with respect to prior training sets. Box and whisker charts depict the distribution of maximum % sequence identity, with respect to a training set, of all TBM/TBM-hard entries in a given ProteinNet test set (CASP set). Comparisons are made for each ProteinNet test set with respect to its corresponding and prior training sets, e.g. for CASP 11 with respect to ProteinNet 7–11 training sets. Color indicates training set used
Similar articles
- Protein contact prediction by integrating deep multiple sequence alignments, coevolution and machine learning.
Adhikari B, Hou J, Cheng J. Adhikari B, et al. Proteins. 2018 Mar;86 Suppl 1(Suppl 1):84-96. doi: 10.1002/prot.25405. Epub 2017 Oct 31. Proteins. 2018. PMID: 29047157 Free PMC article. - SidechainNet: An all-atom protein structure dataset for machine learning.
King JE, Koes DR. King JE, et al. Proteins. 2021 Nov;89(11):1489-1496. doi: 10.1002/prot.26169. Epub 2021 Jul 12. Proteins. 2021. PMID: 34213059 Free PMC article. - Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model.
Wang S, Sun S, Li Z, Zhang R, Xu J. Wang S, et al. PLoS Comput Biol. 2017 Jan 5;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28056090 Free PMC article. - Machine Learning Approaches for Quality Assessment of Protein Structures.
Chen J, Siu SWI. Chen J, et al. Biomolecules. 2020 Apr 17;10(4):626. doi: 10.3390/biom10040626. Biomolecules. 2020. PMID: 32316682 Free PMC article. Review. - A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction.
Moult J. Moult J. Curr Opin Struct Biol. 2005 Jun;15(3):285-9. doi: 10.1016/j.sbi.2005.05.011. Curr Opin Struct Biol. 2005. PMID: 15939584 Review.
Cited by
- Linking Protein Stability to Pathogenicity: Predicting Clinical Significance of Single-Missense Mutations in Ocular Proteins Using Machine Learning.
Majid I, Sergeev YV. Majid I, et al. Int J Mol Sci. 2024 Oct 30;25(21):11649. doi: 10.3390/ijms252111649. Int J Mol Sci. 2024. PMID: 39519200 Free PMC article. - Machine Learning Using Template-Based-Predicted Structure of Haemagglutinin Predicts Pathogenicity of Avian Influenza.
Shin JH, Kim SJ, Kim G, Kim HR, Ko KS. Shin JH, et al. J Microbiol Biotechnol. 2024 Oct 28;34(10):2033-2040. doi: 10.4014/jmb.2405.05022. Epub 2024 Aug 6. J Microbiol Biotechnol. 2024. PMID: 39252651 Free PMC article. - PCP-GC-LM: single-sequence-based protein contact prediction using dual graph convolutional neural network and convolutional neural network.
Ouyang J, Gao Y, Yang Y. Ouyang J, et al. BMC Bioinformatics. 2024 Sep 2;25(1):287. doi: 10.1186/s12859-024-05914-3. BMC Bioinformatics. 2024. PMID: 39223474 Free PMC article. - HERMES: Holographic Equivariant neuRal network model for Mutational Effect and Stability prediction.
Visani GM, Pun MN, Galvin W, Daniel E, Borisiak K, Wagura U, Nourmohammad A. Visani GM, et al. bioRxiv [Preprint]. 2024 Oct 2:2024.07.09.602403. doi: 10.1101/2024.07.09.602403. bioRxiv. 2024. PMID: 39026838 Free PMC article. Preprint. - Evaluating generalizability of artificial intelligence models for molecular datasets.
Ektefaie Y, Shen A, Bykova D, Marin M, Zitnik M, Farhat M. Ektefaie Y, et al. bioRxiv [Preprint]. 2024 Feb 28:2024.02.25.581982. doi: 10.1101/2024.02.25.581982. bioRxiv. 2024. PMID: 38464295 Free PMC article. Preprint.
References
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. - DOI
MeSH terms
Substances
Grants and funding
- P50 GM107618/GM/NIGMS NIH HHS/United States
- U54 CA225088/CA/NCI NIH HHS/United States
- U54-CA225088/National Institutes of Health
- P50-GM107618/National Institute of General Medical Sciences
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous