Eukaryogenesis, a syntrophy affair - PubMed (original) (raw)
Comment
Eukaryogenesis, a syntrophy affair
Purificación López-García et al. Nat Microbiol. 2019 Jul.
Abstract
Eukaryotes evolved from a symbiosis involving alphaproteobacteria and archaea phylogenetically nested within the Asgard clade. Two recent studies explore the metabolic capabilities of Asgard lineages, supporting refined symbiotic metabolic interactions that might have operated at the dawn of eukaryogenesis.
Figures
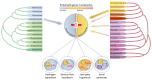
Figure 1. Metabolic symbiosis at the origin of eukaryotes.
Current phylogenomic evidence supports symbiotic models for the origin of the eukaryotic cell. Eukaryotic genomes are mosaics containing a substantial number of genes (~1,000) of archaeal and bacterial ancestry that can now be traced to specific lineages,,. This information supports the idea that eukaryotes evolved from a symbiosis between a member of the recently described Asgard archaea more closely related (so far) to the Heimdallarchaeota and, at least, the facultatively aerobic alphaproteobacterium that gave rise to the mitochondrion. Comparative analyses of the Asgard archaeal metabolic potential allow Spang et al. and Bulzu et al. to conclude that Asgard archaea were primarily organoheterotrophic organisms that can produce and consume hydrogen. Some Heimdallarchaeota also gained the capability to use oxygen and nitrate as final electron acceptors by horizontal gene transfer in a later stage. Based on Asgard archaeal metabolic reconstruction and ecological considerations, Spang et al. propose the ‘reverse flow model’. This refined symbiogenetic model for the origin of eukaryotes invokes a metabolic symbiosis, or syntrophy, mediated by hydrogen or electron transfer between archaea and bacteria. However, unlike the original hydrogen and syntrophy hypotheses, which proposed interspecies hydrogen transfer from the bacterial to the archaeal symbiont, the ‘reverse flow model’ involves electron or hydrogen flow from the archaeal to the bacterial symbiont. This eukaryogenetic syntrophy likely established in anoxic or microoxic environments,. Although the model specifically involves one archaeon and one bacterium, Spang et al. leave open the possibility that symbiotic interactions with other prokaryotes might have intervened, in consonance with recent proposals for serial symbioses.
Comment on
- Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche.
Bulzu PA, Andrei AŞ, Salcher MM, Mehrshad M, Inoue K, Kandori H, Beja O, Ghai R, Banciu HL. Bulzu PA, et al. Nat Microbiol. 2019 Jul;4(7):1129-1137. doi: 10.1038/s41564-019-0404-y. Epub 2019 Apr 1. Nat Microbiol. 2019. PMID: 30936485 - Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism.
Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, Greening C, Baker BJ, Ettema TJG. Spang A, et al. Nat Microbiol. 2019 Jul;4(7):1138-1148. doi: 10.1038/s41564-019-0406-9. Epub 2019 Apr 1. Nat Microbiol. 2019. PMID: 30936488
Similar articles
- The Syntrophy hypothesis for the origin of eukaryotes revisited.
López-García P, Moreira D. López-García P, et al. Nat Microbiol. 2020 May;5(5):655-667. doi: 10.1038/s41564-020-0710-4. Epub 2020 Apr 27. Nat Microbiol. 2020. PMID: 32341569 Review. - The symbiotic origin of the eukaryotic cell.
López-García P, Moreira D. López-García P, et al. C R Biol. 2023 May 30;346:55-73. doi: 10.5802/crbiol.118. C R Biol. 2023. PMID: 37254790 Review. - Open Questions on the Origin of Eukaryotes.
López-García P, Moreira D. López-García P, et al. Trends Ecol Evol. 2015 Nov;30(11):697-708. doi: 10.1016/j.tree.2015.09.005. Epub 2015 Oct 8. Trends Ecol Evol. 2015. PMID: 26455774 Free PMC article. Review. - The changing view of eukaryogenesis - fossils, cells, lineages and how they all come together.
Dacks JB, Field MC, Buick R, Eme L, Gribaldo S, Roger AJ, Brochier-Armanet C, Devos DP. Dacks JB, et al. J Cell Sci. 2016 Oct 15;129(20):3695-3703. doi: 10.1242/jcs.178566. Epub 2016 Sep 26. J Cell Sci. 2016. PMID: 27672020 Review. - Timing the origin of eukaryotic cellular complexity with ancient duplications.
Vosseberg J, van Hooff JJE, Marcet-Houben M, van Vlimmeren A, van Wijk LM, Gabaldón T, Snel B. Vosseberg J, et al. Nat Ecol Evol. 2021 Jan;5(1):92-100. doi: 10.1038/s41559-020-01320-z. Epub 2020 Oct 26. Nat Ecol Evol. 2021. PMID: 33106602 Free PMC article.
Cited by
- A Case Study of Eukaryogenesis: The Evolution of Photoreception by Photolyase/Cryptochrome Proteins.
Miles JA, Davies TA, Hayman RD, Lorenzen G, Taylor J, Anjarwalla M, Allen SJR, Graham JWD, Taylor PC. Miles JA, et al. J Mol Evol. 2020 Nov;88(8-9):662-673. doi: 10.1007/s00239-020-09965-x. Epub 2020 Sep 26. J Mol Evol. 2020. PMID: 32979052 Free PMC article. - The origin and evolution of methanogenesis and Archaea are intertwined.
Mei R, Kaneko M, Imachi H, Nobu MK. Mei R, et al. PNAS Nexus. 2023 Jan 31;2(2):pgad023. doi: 10.1093/pnasnexus/pgad023. eCollection 2023 Feb. PNAS Nexus. 2023. PMID: 36874274 Free PMC article. - Evolution of glutamatergic signaling and synapses.
Moroz LL, Nikitin MA, Poličar PG, Kohn AB, Romanova DY. Moroz LL, et al. Neuropharmacology. 2021 Nov 1;199:108740. doi: 10.1016/j.neuropharm.2021.108740. Epub 2021 Jul 31. Neuropharmacology. 2021. PMID: 34343611 Free PMC article. Review. - Disentangling a metabolic cross-feeding in a halophilic archaea-bacteria consortium.
Medina-Chávez NO, Torres-Cerda A, Chacón JM, Harcombe WR, De la Torre-Zavala S, Travisano M. Medina-Chávez NO, et al. Front Microbiol. 2023 Dec 21;14:1276438. doi: 10.3389/fmicb.2023.1276438. eCollection 2023. Front Microbiol. 2023. PMID: 38179456 Free PMC article. - Unraveling the Metabolic Potential of Asgardarchaeota in a Sediment from the Mediterranean Hydrocarbon-Contaminated Water Basin Mar Piccolo (Taranto, Italy).
Firrincieli A, Negroni A, Zanaroli G, Cappelletti M. Firrincieli A, et al. Microorganisms. 2021 Apr 16;9(4):859. doi: 10.3390/microorganisms9040859. Microorganisms. 2021. PMID: 33923677 Free PMC article.
References
- Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. - PubMed