C-type lectin domain group 14 proteins in vascular biology, cancer and inflammation - PubMed (original) (raw)
Review
. 2019 Sep;286(17):3299-3332.
doi: 10.1111/febs.14985. Epub 2019 Jul 29.
Affiliations
- PMID: 31287944
- PMCID: PMC6852297
- DOI: 10.1111/febs.14985
Review
C-type lectin domain group 14 proteins in vascular biology, cancer and inflammation
Kabir A Khan et al. FEBS J. 2019 Sep.
Abstract
The C-type lectin domain (CTLD) group 14 family of transmembrane glycoproteins consist of thrombomodulin, CD93, CLEC14A and CD248 (endosialin or tumour endothelial marker-1). These cell surface proteins exhibit similar ectodomain architecture and yet mediate a diverse range of cellular functions, including but not restricted to angiogenesis, inflammation and cell adhesion. Thrombomodulin, CD93 and CLEC14A can be expressed by endothelial cells, whereas CD248 is expressed by vasculature associated pericytes, activated fibroblasts and tumour cells among other cell types. In this article, we review the current literature of these family members including their expression profiles, interacting partners, as well as established and speculated functions. We focus primarily on their roles in the vasculature and inflammation as well as their contributions to tumour immunology. The CTLD group 14 family shares several characteristic features including their ability to be proteolytically cleaved and engagement of some shared extracellular matrix ligands. Each family member has strong links to tumour development and in particular CD93, CLEC14A and CD248 have been proposed as attractive candidate targets for cancer therapy.
Keywords: C-type lectin; CD248; CD93; CLEC14A; cancer; extracellular matrix; group XIV; immuno-oncology; thrombomodulin; vascular targeting.
© 2019 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
KAK and RB are inventors on patents WO/2016/116760 entitled ‘Inhibitors of the interaction between CLEC14A and Multimerin‐2 for inhibition of angiogenesis’ and the related filed patents GB1612860.5, GB1612534.6 and GB1702926.5.
Figures
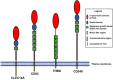
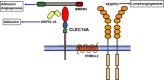
Figure 1
CTLD
group 14 family proteins. Schematic diagrams of the
CTLD
group 14 family proteins. Each protein is drawn to relative scale based on primary amino acid sequence length. The
CTLD
is shown in red, the sushi in blue and the
EGF
repeats in green.
Figure 2
Alignments of
CTLD
group 14 family members based on amino acid sequence. Amino acid alignments of the whole primary sequence of each human family member using PRALINE 229. The following protein sequences were used thrombomodulin (
http://www.uniprot.org/uniprot/P07204
),
CD
93 (
http://www.uniprot.org/uniprot/Q9NPY3
),
CLEC
14A (
http://www.uniprot.org/uniprot/Q86T13
) and
CD
248 (
http://www.uniprot.org/uniprot/Q9HCU0
).
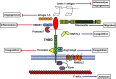
Figure 3
Schematic of thrombomodulin protein structure with ligand binding partners. Thrombomodulin
CTLD
has been shown to interact with fibronectin,
HMGB
1, Kringle 1–5, Lewis Y antigen and
HSP
70‐1. The
CTLD
may be proteolytically cleaved by an as yet unidentified
MMP
. Thrombin binds to the 5th and 6th
EGF
domains, this binding is in competition with Ang1 and/or Ang2.
RHBDL
2 can cleave the whole ECD of thrombomodulin as can neutrophil elastase, cathepsin G and proteinase 3. The cytoplasmic tail binds to ezrin which in turn links thrombomodulin to the actin cytoskeleton.
Figure 4
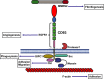
Schematic of
CD
248 structure with ligand binding partners.
CD
248
CTLD
binds to fibronectin, Mac‐2
BP
, Collagens I and
IV
and
MMRN
2. There are currently no known direct intracellular interaction partners for
CD
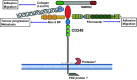
Figure 5
Schematic of
CD
93 structure with ligand binding partners.
CD
93
CTLD
binds to
MMRN
2. The whole ECD has been shown to be cleaved by an as yet unidentified metalloproteinase. The intracellular cytoplasmic domain binds to moesin which in turn links
CD
93 to the actin cytoskeleton. The cytoplasmic domain also binds to Cbl and
GIPC
1 and src.
Figure 6
Schematic of
CLEC
14A protein with ligand binding partners.
CLEC
14A
CTLD
binds to
MMRN
2 and to
HSP
70‐1A. The whole ECD can be cleaved by
RHBDL
2. There are currently no known direct intracellular partners for
CLEC
14A.
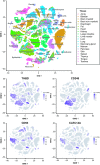
Figure 7
Expression of
CTLD
group 14 family members in mouse tissues. The Tabula Muris database was used to determine which mouse cell types expressed each
CTLD
group 14 family gene from data acquired through fluorescence activated cell sorting and single‐cell gene expression analysis. The t‐
SNE
plot at the top displays annotations of each cell type and shows a legend of colours corresponding to which organ or tissue type that cell was from. The lower t‐
SNE
plots display in which cell types each family member was expressed (purple), ln(1 +
CPM
) is the natural logarithm of counts per million + 1.
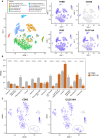
Figure 8
Endothelial expression of
CTLD
group 14 family members in mouse tissues. (A) The Tabula Muris database was used to create t‐
SNE
plots of all endothelial cells from different organs as well as brain pericytes. The t‐
SNE
plot at the top left displays a legend of colours corresponding to which organ or tissue type that cell was from. Expression of each
CTLD
group 14 family member within these cell types are displayed as t‐
SNE
plots. (B) Single‐cell sequencing data analysed as fragments per kilobase million was used to compare
CD
93 and
CLEC
14A expression in different endothelial cells from different organs. Wilcoxon statistical test was used to compare ****P ≤ 0.0001 Aorta
EC
s n = 262, Brain nonmyeloid
EC
n = 1250, Diaphragm
EC
n = 154, Fat
EC
n = 1180, Heart
EC
n = 2274, Heart endocardial cell n = 350, Kidney
EC
n = 80, Limb Muscle
EC
n = 258, Liver
EC
n = 392, Lung
EC
n = 1476, Mammary gland
EC
n = 98, Pancreas
EC
n = 98, Trachea
EC
n = 66. (C) t‐
SNE
plots of lung endothelium alone were created which revealed the presence of a cluster of cells expressing low levels of
CD
93 when compared with all other lung endothelial cells but similar levels of
CLEC
14A (grey ellipse).
Similar articles
- Multimerin-2 is a ligand for group 14 family C-type lectins CLEC14A, CD93 and CD248 spanning the endothelial pericyte interface.
Khan KA, Naylor AJ, Khan A, Noy PJ, Mambretti M, Lodhia P, Athwal J, Korzystka A, Buckley CD, Willcox BE, Mohammed F, Bicknell R. Khan KA, et al. Oncogene. 2017 Nov 2;36(44):6097-6108. doi: 10.1038/onc.2017.214. Epub 2017 Jul 3. Oncogene. 2017. PMID: 28671670 Free PMC article. - Soluble expression of disulfide-bonded C-type lectin like domain of human CD93 in the cytoplasm of Escherichia coli.
Nativel B, Figuester A, Andries J, Planesse C, Couprie J, Gasque P, Viranaicken W, Iwema T. Nativel B, et al. J Immunol Methods. 2016 Dec;439:67-73. doi: 10.1016/j.jim.2016.10.003. Epub 2016 Oct 12. J Immunol Methods. 2016. PMID: 27742562 - Inflammatory Mesenchymal Stem Cells Express Abundant Membrane-Bound and Soluble Forms of C-Type Lectin-like CD248.
Payet M, Ah-Pine F, Guillot X, Gasque P. Payet M, et al. Int J Mol Sci. 2023 May 31;24(11):9546. doi: 10.3390/ijms24119546. Int J Mol Sci. 2023. PMID: 37298499 Free PMC article. - Group XIV C-type lectins: emerging targets in tumor angiogenesis.
Yee EJ, Vigil I, Sun Y, Torphy RJ, Schulick RD, Zhu Y. Yee EJ, et al. Angiogenesis. 2024 May;27(2):173-192. doi: 10.1007/s10456-024-09907-x. Epub 2024 Mar 12. Angiogenesis. 2024. PMID: 38468017 Free PMC article. Review. - CD93 and related family members: their role in innate immunity.
Greenlee MC, Sullivan SA, Bohlson SS. Greenlee MC, et al. Curr Drug Targets. 2008 Feb;9(2):130-8. doi: 10.2174/138945008783502421. Curr Drug Targets. 2008. PMID: 18288964 Review.
Cited by
- Unveiling the multifaceted role of toll-like receptors in immunity of aquatic animals: pioneering strategies for disease management.
Ghani MU, Chen J, Khosravi Z, Wu Q, Liu Y, Zhou J, Zhong L, Cui H. Ghani MU, et al. Front Immunol. 2024 Oct 17;15:1378111. doi: 10.3389/fimmu.2024.1378111. eCollection 2024. Front Immunol. 2024. PMID: 39483482 Free PMC article. Review. - Type-H endothelial cell protein Clec14a orchestrates osteoblast activity during trabecular bone formation and patterning.
Neag G, Lewis J, Turner JD, Manning JE, Dean I, Finlay M, Poologasundarampillai G, Woods J, Sahu MA, Khan KA, Begum J, McGettrick HM, Bellantuono I, Heath V, Jones SW, Buckley CD, Bicknell R, Naylor AJ. Neag G, et al. Commun Biol. 2024 Oct 11;7(1):1296. doi: 10.1038/s42003-024-06971-3. Commun Biol. 2024. PMID: 39394430 Free PMC article. - Clinicopathological association of CD93 expression in gastric adenocarcinoma.
Shen Y, Wu Y, Hao M, Fu M, Zhu K, Luo P, Wang J. Shen Y, et al. J Cancer Res Clin Oncol. 2024 Aug 27;150(8):400. doi: 10.1007/s00432-024-05874-4. J Cancer Res Clin Oncol. 2024. PMID: 39190192 Free PMC article. - Regulation of matrix reloading by tumor endothelial marker 1 protects against abdominal aortic aneurysm.
Hong YK, Cheng TL, Hsu CK, Lee FT, Chang BI, Wang KC, Chang LY, Wu HL, Lai CH. Hong YK, et al. Int J Biol Sci. 2024 Jul 2;20(10):3691-3709. doi: 10.7150/ijbs.93526. eCollection 2024. Int J Biol Sci. 2024. PMID: 39113704 Free PMC article. - Recent Insights into Cellular and Molecular Mechanisms of Defective Angiogenesis in Systemic Sclerosis.
Romano E, Rosa I, Fioretto BS, Manetti M. Romano E, et al. Biomedicines. 2024 Jun 14;12(6):1331. doi: 10.3390/biomedicines12061331. Biomedicines. 2024. PMID: 38927538 Free PMC article. Review.
References
- Zelensky AN & Gready JE (2005) The C‐type lectin‐like domain superfamily. FEBS J 272, 6179–6217. - PubMed
- McMahon SA, Miller JL, Lawton JA, Kerkow DE, Hodes A, Marti‐Renom MA, Doulatov S, Narayanan E, Sali A, Miller JF et al (2005) The C‐type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol 12, 886–892. - PubMed
- Norman DG, Barlow PN, Baron M, Day AJ, Sim RB & Campbell ID (1991) Three‐dimensional structure of a complement control protein module in solution. J Mol Biol 219, 717–725. - PubMed
- Wei X‐Q, Orchardson M, Gracie JA, Leung BP, Gao BM, Guan H, Niedbala W, Paterson GK, McInnes IB & Liew FY (2001) The sushi domain of soluble IL‐15 receptor is essential for binding IL‐15 and inhibiting inflammatory and allogenic responses in vitro and in vivo. J Immunol 167, 277–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous