Scalable and accurate deep learning with electronic health records - PubMed (original) (raw)
doi: 10.1038/s41746-018-0029-1. eCollection 2018.
Eyal Oren # 1, Kai Chen 1, Andrew M Dai 1, Nissan Hajaj 1, Michaela Hardt 1, Peter J Liu 1, Xiaobing Liu 1, Jake Marcus 1, Mimi Sun 1, Patrik Sundberg 1, Hector Yee 1, Kun Zhang 1, Yi Zhang 1, Gerardo Flores 1, Gavin E Duggan 1, Jamie Irvine 1, Quoc Le 1, Kurt Litsch 1, Alexander Mossin 1, Justin Tansuwan 1, De Wang 1, James Wexler 1, Jimbo Wilson 1, Dana Ludwig 2, Samuel L Volchenboum 3, Katherine Chou 1, Michael Pearson 1, Srinivasan Madabushi 1, Nigam H Shah 4, Atul J Butte 2, Michael D Howell 1, Claire Cui 1, Greg S Corrado 1, Jeffrey Dean 1
Affiliations
- PMID: 31304302
- PMCID: PMC6550175
- DOI: 10.1038/s41746-018-0029-1
Scalable and accurate deep learning with electronic health records
Alvin Rajkomar et al. NPJ Digit Med. 2018.
Abstract
Predictive modeling with electronic health record (EHR) data is anticipated to drive personalized medicine and improve healthcare quality. Constructing predictive statistical models typically requires extraction of curated predictor variables from normalized EHR data, a labor-intensive process that discards the vast majority of information in each patient's record. We propose a representation of patients' entire raw EHR records based on the Fast Healthcare Interoperability Resources (FHIR) format. We demonstrate that deep learning methods using this representation are capable of accurately predicting multiple medical events from multiple centers without site-specific data harmonization. We validated our approach using de-identified EHR data from two US academic medical centers with 216,221 adult patients hospitalized for at least 24 h. In the sequential format we propose, this volume of EHR data unrolled into a total of 46,864,534,945 data points, including clinical notes. Deep learning models achieved high accuracy for tasks such as predicting: in-hospital mortality (area under the receiver operator curve [AUROC] across sites 0.93-0.94), 30-day unplanned readmission (AUROC 0.75-0.76), prolonged length of stay (AUROC 0.85-0.86), and all of a patient's final discharge diagnoses (frequency-weighted AUROC 0.90). These models outperformed traditional, clinically-used predictive models in all cases. We believe that this approach can be used to create accurate and scalable predictions for a variety of clinical scenarios. In a case study of a particular prediction, we demonstrate that neural networks can be used to identify relevant information from the patient's chart.
Keywords: Machine learning; Medical research.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
Fig. 1
This boxplot displays the amount of data (on a log scale) in the EHR, along with its temporal variation across the course of an admission. We define a token as a single data element in the electronic health record, like a medication name, at a specific point in time. Each token is considered as a potential predictor by the deep learning model. The line within the boxplot represents the median, the box represents the interquartile range (IQR), and the whiskers are 1.5 times the IQR. The number of tokens increased steadily from admission to discharge. At discharge, the median number of tokens for Hospital A was 86,477 and for Hospital B was 122,961
Fig. 2
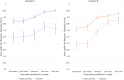
The area under the receiver operating characteristic curves are shown for predictions of inpatient mortality made by deep learning and baseline models at 12 h increments before and after hospital admission. For inpatient mortality, the deep learning model achieves higher discrimination at every prediction time compared to the baseline for both the University of California, San Francisco (UCSF) and University of Chicago Medicine (UCM) cohorts. Both models improve in the first 24 h, but the deep learning model achieves a similar level of accuracy approximately 24 h earlier for UCM and even 48 h earlier for UCSF. The error bars represent the bootstrapped 95% confidence interval
Fig. 3
The patient record shows a woman with metastatic breast cancer with malignant pleural effusions and empyema. The patient timeline at the top of the figure contains circles for every time-step for which at least a single token exists for the patient, and the horizontal lines show the data type. There is a close-up view of the most recent data points immediately preceding a prediction made 24 h after admission. We trained models for each data type and highlighted in red the tokens which the models attended to—the non-highlighted text was not attended to but is shown for context. The models pick up features in the medications, nursing flowsheets, and clinical notes relevant to the prediction
Fig. 4
Data from each health system were mapped to an appropriate FHIR (Fast Healthcare Interoperability Resources) resource and placed in temporal order. This conversion did not harmonize or standardize the data from each health system other than map them to the appropriate resource. The deep learning model could use all data available prior to the point when the prediction was made. Therefore, each prediction, regardless of the task, used the same data
Similar articles
- Developing a FHIR-based EHR phenotyping framework: A case study for identification of patients with obesity and multiple comorbidities from discharge summaries.
Hong N, Wen A, Stone DJ, Tsuji S, Kingsbury PR, Rasmussen LV, Pacheco JA, Adekkanattu P, Wang F, Luo Y, Pathak J, Liu H, Jiang G. Hong N, et al. J Biomed Inform. 2019 Nov;99:103310. doi: 10.1016/j.jbi.2019.103310. Epub 2019 Oct 14. J Biomed Inform. 2019. PMID: 31622801 Free PMC article. - Predicting next-day discharge via electronic health record access logs.
Zhang X, Yan C, Malin BA, Patel MB, Chen Y. Zhang X, et al. J Am Med Inform Assoc. 2021 Nov 25;28(12):2670-2680. doi: 10.1093/jamia/ocab211. J Am Med Inform Assoc. 2021. PMID: 34592753 Free PMC article. - Early Detection of Septic Shock Onset Using Interpretable Machine Learners.
Misra D, Avula V, Wolk DM, Farag HA, Li J, Mehta YB, Sandhu R, Karunakaran B, Kethireddy S, Zand R, Abedi V. Misra D, et al. J Clin Med. 2021 Jan 15;10(2):301. doi: 10.3390/jcm10020301. J Clin Med. 2021. PMID: 33467539 Free PMC article. - Using Predictive Models to Improve Care for Patients Hospitalized with COVID-19 [Internet].
Kaushal R, Zhang Y, Banerjee S, Weiner M, Su C, Wang F, Schenck E, Goyal P, Khullar D, Steel P, Flory J, Hupert N, Schpero W, Díaz I, Choi J, Wu Y, Orlander D, Morozyuk D. Kaushal R, et al. Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2023 Jan. Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2023 Jan. PMID: 38976624 Free Books & Documents. Review. - Electronic Health Record and Semantic Issues Using Fast Healthcare Interoperability Resources: Systematic Mapping Review.
Amar F, April A, Abran A. Amar F, et al. J Med Internet Res. 2024 Jan 30;26:e45209. doi: 10.2196/45209. J Med Internet Res. 2024. PMID: 38289660 Free PMC article. Review.
Cited by
- Survival models and longitudinal medical events for hospital readmission forecasting.
Davis S, Greiner R. Davis S, et al. BMC Health Serv Res. 2024 Nov 13;24(1):1394. doi: 10.1186/s12913-024-11771-w. BMC Health Serv Res. 2024. PMID: 39538197 Free PMC article. - Multisource representation learning for pediatric knowledge extraction from electronic health records.
Li M, Li X, Pan K, Geva A, Yang D, Sweet SM, Bonzel CL, Ayakulangara Panickan V, Xiong X, Mandl K, Cai T. Li M, et al. NPJ Digit Med. 2024 Nov 13;7(1):319. doi: 10.1038/s41746-024-01320-4. NPJ Digit Med. 2024. PMID: 39533050 Free PMC article. - Early prediction of hypertensive disorders of pregnancy toward preventive early intervention.
Mizuno S, Nagaie S, Sugawara J, Tamiya G, Obara T, Ishikuro M, Kuriyama S, Yaegashi N, Tanaka H, Yamamoto M, Ogishima S. Mizuno S, et al. AJOG Glob Rep. 2024 Jul 27;4(4):100383. doi: 10.1016/j.xagr.2024.100383. eCollection 2024 Nov. AJOG Glob Rep. 2024. PMID: 39524694 Free PMC article. - Ten challenges and opportunities in computational immuno-oncology.
Bao R, Hutson A, Madabhushi A, Jonsson VD, Rosario SR, Barnholtz-Sloan JS, Fertig EJ, Marathe H, Harris L, Altreuter J, Chen Q, Dignam J, Gentles AJ, Gonzalez-Kozlova E, Gnjatic S, Kim E, Long M, Morgan M, Ruppin E, Valen DV, Zhang H, Vokes N, Meerzaman D, Liu S, Van Allen EM, Xing Y. Bao R, et al. J Immunother Cancer. 2024 Oct 26;12(10):e009721. doi: 10.1136/jitc-2024-009721. J Immunother Cancer. 2024. PMID: 39461879 Free PMC article. Review. - Applicability of the adjusted morbidity groups algorithm for healthcare programming: results of a pilot study in Italy.
Papa R, Balducci F, Franceschini G, Pompili M, De Marco M, Roca J, González-Colom R, Monterde D. Papa R, et al. BMC Public Health. 2024 Oct 17;24(1):2869. doi: 10.1186/s12889-024-20398-9. BMC Public Health. 2024. PMID: 39420326 Free PMC article.
References
- The Digital Universe: Driving Data Growth in Healthcare. Available at: https://www.emc.com/analyst-report/digital-universe-healthcare-vertical-... (Accessed 23 Feb 2017).
LinkOut - more resources
Full Text Sources
Other Literature Sources