Tumor mutational burden quantification from targeted gene panels: major advancements and challenges - PubMed (original) (raw)
Review
Tumor mutational burden quantification from targeted gene panels: major advancements and challenges
Laura Fancello et al. J Immunother Cancer. 2019.
Abstract
Tumor mutational burden (TMB), the total number of somatic coding mutations in a tumor, is emerging as a promising biomarker for immunotherapy response in cancer patients. TMB can be quantitated by a number of NGS-based sequencing technologies. Whole Exome Sequencing (WES) allows comprehensive measurement of TMB and is considered the gold standard. However, to date WES remains confined to research settings, due to high cost of the large genomic space sequenced. In the clinical setting, instead, targeted enrichment panels (gene panels) of various genomic sizes are emerging as the routine technology for TMB assessment. This stimulated the development of various methods for panel-based TMB quantification, and prompted the multiplication of studies assessing whether TMB can be confidently estimated from the smaller genomic space sampled by gene panels. In this review, we inventory the collection of available gene panels tested for this purpose, illustrating their technical specifications and describing their accuracy and clinical value in TMB assessment. Moreover, we highlight how various experimental, platform-related or methodological variables, as well as bioinformatic pipelines, influence panel-based TMB quantification. The lack of harmonization in panel-based TMB quantification, of adequate methods to convert TMB estimates across different panels and of robust predictive cutoffs, currently represents one of the main limitations to adopt TMB as a biomarker in clinical practice. This overview on the heterogeneous landscape of panel-based TMB quantification aims at providing a context to discuss common standards and illustrates the strong need of further validation and consolidation studies for the clinical interpretation of panel-based TMB values.
Keywords: Gene panels; Immunotherapy; TMB; Targeted enrichment sequencing; Tumor mutational burden.
Conflict of interest statement
LF, SG, PGP, and LM declare that they have no competing interest.
Figures
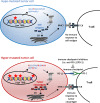
Fig. 1
Tumor mutational burden as immunotherapy biomarker. Interaction between tumor mutational burden, neoantigen production and immune checkpoints. Hyper-mutated tumors (bottom) are more likely than hypo-mutated tumors (top) to generate tumor-specific peptides (neoantigens) recognized by the immune system. However, immune surveillance can be restrained by simultaneous high expression of PD-L1, which delivers a suppressive signal to T cells. PD-L1/PD-1 interaction and other immune checkpoints can be inhibited by immune checkpoint inhibitors, restoring immune response
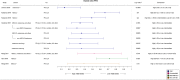
Fig. 2
TMB association with progression-free survival. Forest plot of hazard ratios (HR) comparing progression-free survival (PFS) between patients with high or low TMB, as indicated in the “Comparison” column. If not specified otherwise, TMB is reported as number of mutations per Mb. All patients were treated with immune checkpoint inhibitors (ICI). Bars represent the 95% confidence intervals. Size of the box is proportional to precision. Reference to the study and the analyzed cancer type are also reported together with the log-rank _p_-value. Q1-Q4: quartiles; VUS: variants of unknown significance. *: TMB quantified from blood; **: Cox proportional hazards model adjusted for age, gender, disease stage and prior therapy by ipilimumab
Fig. 3
Differences in the workflow for panel-based TMB quantification. a. Overview of the factors influencing panel-based TMB quantification. Several variables in library construction, sequencing and in the pipeline to call mutations influence panel-based TMB quantification. Furthermore, panel-based TMB quantification is influenced by differences in the bioinformatic method to extrapolate global TMB from mutations identified in the narrow genomic region targeted by the gene panel. b. Differences across various studies in panel-based TMB quantification: gene panel technical specifications, preanalytical factors and the bioinformatics workflow used to extrapolate from the genomic space targeted by gene panels global TMB are described. FM1: Foundation Medicine’s FoundationOne panel (v1: 185 genes, v2: 236 genes, v3: 315 genes, v4: 405 genes); NA: not available; ±: algorithm developed by Sun et al. for in silico removal of germline variants [74]
References
- Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Ri H, et al. Durvalumab after chemoradiotherapy in stage III non-small cell lung cancer. N Engl J Med. 2017;377:1919–1929. - PubMed
- Rosenberg Jonathan E, Hoffman-Censits Jean, Powles Tom, van der Heijden Michiel S, Balar Arjun V, Necchi Andrea, Dawson Nancy, O'Donnell Peter H, Balmanoukian Ani, Loriot Yohann, Srinivas Sandy, Retz Margitta M, Grivas Petros, Joseph Richard W, Galsky Matthew D, Fleming Mark T, Petrylak Daniel P, Perez-Gracia Jose Luis, Burris Howard A, Castellano Daniel, Canil Christina, Bellmunt Joaquim, Bajorin Dean, Nickles Dorothee, Bourgon Richard, Frampton Garrett M, Cui Na, Mariathasan Sanjeev, Abidoye Oyewale, Fine Gregg D, Dreicer Robert. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. The Lancet. 2016;387(10031):1909–1920. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources