GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network - PubMed (original) (raw)
doi: 10.1186/s12916-019-1364-z.
Todd Lingren 3 4, Yongbo Huang 5, Sreeja Parameswaran 5, Beth L Cobb 5, Ian B Stanaway 6, John J Connolly 7, Frank D Mentch 7, Barbara Benoit 8, Xinnan Niu 9, Wei-Qi Wei 9, Robert J Carroll 9, Jennifer A Pacheco 10, Isaac T W Harley 11, Senad Divanovic 11, David S Carrell 12, Eric B Larson 12, David J Carey 13, Shefali Verma 14, Marylyn D Ritchie 14, Ali G Gharavi 15, Shawn Murphy 16, Marc S Williams 17, David R Crosslin 6, Gail P Jarvik 18, Iftikhar J Kullo 19, Hakon Hakonarson 7 20, Rongling Li 21; eMERGE Network; Stavra A Xanthakos 22, John B Harley 5 3 23
Affiliations
- PMID: 31311600
- PMCID: PMC6636057
- DOI: 10.1186/s12916-019-1364-z
GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network
Bahram Namjou et al. BMC Med. 2019.
Abstract
Background: Non-alcoholic fatty liver disease (NAFLD) is a common chronic liver illness with a genetically heterogeneous background that can be accompanied by considerable morbidity and attendant health care costs. The pathogenesis and progression of NAFLD is complex with many unanswered questions. We conducted genome-wide association studies (GWASs) using both adult and pediatric participants from the Electronic Medical Records and Genomics (eMERGE) Network to identify novel genetic contributors to this condition.
Methods: First, a natural language processing (NLP) algorithm was developed, tested, and deployed at each site to identify 1106 NAFLD cases and 8571 controls and histological data from liver tissue in 235 available participants. These include 1242 pediatric participants (396 cases, 846 controls). The algorithm included billing codes, text queries, laboratory values, and medication records. Next, GWASs were performed on NAFLD cases and controls and case-only analyses using histologic scores and liver function tests adjusting for age, sex, site, ancestry, PC, and body mass index (BMI).
Results: Consistent with previous results, a robust association was detected for the PNPLA3 gene cluster in participants with European ancestry. At the PNPLA3-SAMM50 region, three SNPs, rs738409, rs738408, and rs3747207, showed strongest association (best SNP rs738409 p = 1.70 × 10- 20). This effect was consistent in both pediatric (p = 9.92 × 10- 6) and adult (p = 9.73 × 10- 15) cohorts. Additionally, this variant was also associated with disease severity and NAFLD Activity Score (NAS) (p = 3.94 × 10- 8, beta = 0.85). PheWAS analysis link this locus to a spectrum of liver diseases beyond NAFLD with a novel negative correlation with gout (p = 1.09 × 10- 4). We also identified novel loci for NAFLD disease severity, including one novel locus for NAS score near IL17RA (rs5748926, p = 3.80 × 10- 8), and another near ZFP90-CDH1 for fibrosis (rs698718, p = 2.74 × 10- 11). Post-GWAS and gene-based analyses identified more than 300 genes that were used for functional and pathway enrichment analyses.
Conclusions: In summary, this study demonstrates clear confirmation of a previously described NAFLD risk locus and several novel associations. Further collaborative studies including an ethnically diverse population with well-characterized liver histologic features of NAFLD are needed to further validate the novel findings.
Keywords: Fatty liver; GWAS; Genetic polymorphism; NAFLD; PheWAS; Polygenic risk score.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Fig. 1
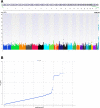
a, b Manhattan plot (a) and Q–Q plot (b) of genome-wide markers for NAFLD in European ancestry (1106 cases and 8571 controls). A total of 1106 cases of NAFLD and 8571 controls were analyzed after quality control. Logistic regression analysis was performed for 7,261,527 variants with MAF > 1% assuming an additive genetic model, adjusted for age, sex, BMI, genotyping site, and genetic ancestry (principal components 1 through 3). Results are plotted as –log10 p values on the _y_-axis by position in chromosome (_x_-axis) (NCBI build 37)
Fig. 2
a–c LocusZoom plot of the associations signals in three previously known regions for NAFLD. a Confirmation at 22q13 for PNPLA3. SNP rs738409 is a missense variation (I148M) in PNPLA3 produced the best effect (p = 1.70 × 10− 20). b Detected signal at 19p12 (GATAD2A, NCAN, TM6SF2) region. The best marker in this study was rs56408111 (p = 5.26 × 10− 6). The linkage disequilibrium (LD) between rs56408111 and previously known SNP rs4808199 was _r_2 = 0.24, _D_’ = 0.74. c Detected signal at 8q24 (TRIB1) genetic region. The best marker in this study (rs2980888) is shown (see also Additional file 1: Table S2). Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant variant are color-coded to reflect their LD with the index SNP (taken from pairwise r2 values from the HapMap CEU database,
). Regional plots were generated using LocusZoom (
http://csg.sph.umich.edu/locuszoom
)
Fig. 3
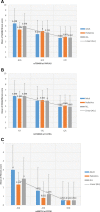
a Means and standard deviations of NAS and fibrosis score (0–12) stratified by genotype of rs738409 at PNPLA3 in 235 NAFLD cases. The results are plotted as the sum of NAS and fibrosis score (0–12) (_y_-axis) against the three genotypes of rs738409 C>G polymorphism (_x_-axis). The results are further sub-divided by age groups (pediatrics, adult, and all). Results for IL17RA (b) and ZFP90 (c) also are shown
Fig. 4
a–d Regional association plots of best effects in case-only linear regression analyses for continuous traits of NAS score, fibrosis, and ALT liver enzyme, respectively. a The best observed effect near the IL17RA region for NAS score. b The most significant effects at 16q22 near ZFP90 gene for fibrosis. c The effect near FABP1 locus for fibrosis. d An effect at 2p22 near XDH for AST liver enzyme
Fig. 5
NAFLD case-control gene-based results using MAGMA as a base and tissue-specific gene expression (GTEx v7 with 30 general tissue types) as a source produced specific enrichment in liver (see “Methods”). List of all MAGMA gene-based results (P < 0.05) is shown in Additional file 1: Table S5
Fig. 6
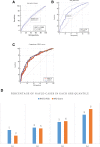
a–d ROC graphical plot that illustrates the diagnostic ability of the binary classifier NAFLD (cases and controls) and NAS score (above and below 5) using weighted GRS score of ten previously published SNPs (GRS-10, see “Results”). The sensitivity and specificity and AUC measures for each plot are also shown. a ROC curve for NAFLD-1106 cases and 8571 controls. b ROC curve for NAS score (79 cases above NAS score ≥ 5 versus 156 controls with score < 5). c Adding SNP rs5748926 near IL17RA improved the ROC curves for NAS score (GRS_11); difference between areas 0.035 (SE = 0.012, p = 0.004). d Distribution of quantiles of weighted 10-SNP GRS in NAFLD (cases and controls) and NAS score (above and below 5); percentage of NAFLD risk increases by increasing GRS quantiles; for NAFLD (cases and controls) from 17% in Q1 to 36% in Q4 (OR = 2.16, 95% CI = 1.81–2.58, p < 0.0001); for NAS score above 5 (defined as case) from 10% in Q1 to 43% in Q4 (OR = 8.50, 95% CI 3.45–20.96). The weighted 10-SNP GRS was calculated by multiplying the sum of the number of risk alleles (0, 1, 2) with the allele-specific effect sizes (beta coefficients) obtained from previous publications (see “Methods”)
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 HG006828/HG/NHGRI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- U01 AI130830/AI/NIAID NIH HHS/United States
- U01 HG006380/HG/NHGRI NIH HHS/United States
- U01 HG008664/HG/NHGRI NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- R01 DK099222/DK/NIDDK NIH HHS/United States
- U01 HG008679/HG/NHGRI NIH HHS/United States
- U01 HG006830/HG/NHGRI NIH HHS/United States
- U01 HG006385/HG/NHGRI NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- R01 HL133786/HL/NHLBI NIH HHS/United States
- P30 AR070549/AR/NIAMS NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- T32 AR007534/AR/NIAMS NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- I01 BX001834/BX/BLRD VA/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
- U01 HG008680/HG/NHGRI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous