Liquid-crystalline phase transitions in lipid droplets are related to cellular states and specific organelle association - PubMed (original) (raw)
Liquid-crystalline phase transitions in lipid droplets are related to cellular states and specific organelle association
Julia Mahamid et al. Proc Natl Acad Sci U S A. 2019.
Abstract
Lipid droplets (LDs) are ubiquitous organelles comprising a central hub for cellular lipid metabolism and trafficking. This role is tightly associated with their interactions with several cellular organelles. Here, we provide a systematic and quantitative structural description of LDs in their native state in HeLa cells enabled by cellular cryoelectron microscopy. LDs consist of a hydrophobic neutral lipid mixture of triacylglycerols (TAG) and cholesteryl esters (CE), surrounded by a single monolayer of phospholipids. We show that under normal culture conditions, LDs are amorphous and that they transition into a smectic liquid-crystalline phase surrounding an amorphous core at physiological temperature under certain cell-cycle stages or metabolic scenarios. Following determination of the crystal lattice spacing of 3.5 nm and of a phase transition temperature below 43 °C, we attributed the liquid-crystalline phase to CE. We suggest that under mitotic arrest and starvation, relative CE levels increase, presumably due to the consumption of TAG metabolites for membrane synthesis and mitochondrial respiration, respectively, supported by direct visualization of LD-mitochondrial membrane contact sites. We hypothesize that the structural phase transition may have a major impact on the accessibility of lipids in LDs to enzymes or lipid transporters. These may become restricted in the smectic phase, affecting the exchange rate of lipids with surrounding membranes and lead to a different surface occupancy of LD-associated proteins. Therefore, the composition and the resulting internal structure of LDs is expected to play a key role in their function as hubs of cellular lipid flux.
Keywords: cholesteryl ester; correlative light and electron microscopy; cryoelectron tomography; cryofocused ion beam; membrane contact sites.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
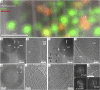
Fig. 1.
LDs exhibit an internal structure of concentric rings in mitotically arrested cells. (A) Overlay of cryo-TEM montage of FIB lamella with a computer-generated oblique slice through a confocal fluorescence microscopy volume (enlarged from yellow frame in
SI Appendix, Fig. S2_G_
.) Green, general neutral lipid dye. Red, fatty acids dye. LDs stained in green or red are distinguishable within the lamella. (B) Enlarged view of frame in A shows two dense LDs in close proximity to a mitochondrion (mito). (C) Enlarged view of frame in B shows on the right the two membranes of the mitochondrion (inner, IMM and outer, OMM), each consisting of a lipid bilayer. The lipid droplet on the left shows a layered structure at its periphery. Granulation observed in the image is due to surface sputter coating with platinum aimed at increasing the conductivity of lamellae for imaging with electrons. (D) Zoom-in of blue frame in
SI Appendix, Fig. S2_H_
shows an LD delineated by ER-membranes and a multilamellar body (MLB). (E) LD is enlarged: the periphery of the LD exhibits a layered organization. A Fourier transform of the image in E' exhibits a single peak of 3.5 nm. (F) Enlarged view of MLB in D shows membrane bilayers. A Fourier transform of the image in F' exhibits several peaks, prominently 4 to 4.9 nm of the lipid bilayer spacing and that of 11 nm corresponding to the space between the membranes. (G) Enlarged view of the center of LD in D (white arrowhead) shows a crystalline lattice (four lattice lines are marked with dotted lines) within a ∼50-nm domain (double-sided arrow delineates the width of the domain). A Fourier transform of the image in G' exhibits a single sharp peak of 3.6 nm. (H) Tomographic slice from a different FIB lamella showing a dense LD delineated by ER membranes. (I) Enlarged view of frame in H shows the layered structure at the periphery of the LD. Spacing between layers is directly measured to be 3.5 nm.
Fig. 2.
LDs exhibit varying internal structure under different cellular states. Top row shows entire LD with Insets representing the fast Fourier transforms (FFT). Bottom rows provide a zoomed-in view at a central slice through the LDs (periphery-center). (Scale bars, 100 nm.) LDs in normally cycling cultures (control) are amorphous, and the FFT only exhibits modulations associated with EM images acquired at defocus. Mitotically arrested cells show a uniform concentric layered structure at their periphery (double-sided arrow), represented by a ring in the FFT. Starved cells also exhibit a layered structure (double-sided arrow), as well as crystalline nanodomains at the center (arrowheads), represented by bright peaks superimposed on a ring of the same spacing in the FFT. Starved cells exposed to heat shock of 43 °C lose the crystalline structure and are amorphous. LDs in Arsenite-treated cells (30′ As) are distorted and exhibit bilayer-like structure at the periphery. The FFT was generated from the framed area.
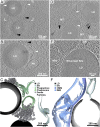
Fig. 3.
Cellular tomography of LDs reveals spatial association with organelles. (A) LDs in a mitotically arrested cell, spatially associated with elongated membranes (white arrowheads) and with 20- to 30-nm-diameter high-density particles (black arrowheads). (B) Enlarged view of frame in A at a different tomographic slice showing that the elongated membranes resemble the classical morphology of phagophores. A closed autophagosome is in close proximity (PH). (C) Annotated 3D segmentation of LDs, putative phagophores/autophagosomes, and high-density particles. (D) Two LDs in a different mitotically arrested cell, associated with the appearance of elongated membranes (white arrowheads) and high-density particles (black arrowheads). Elongated membrane structures with density present within their lumen are annotated by gray arrowheads. Microtubules (MT) are present at high abundance due to the mitotic cell-cycle stage. (E) LD in a starved cell associated with a mitochondrion (Mito) through a tight membrane contact site (arrowhead). An ER membrane is in direct physical contact with the LD. (F) Annotated 3D segmentation of LD contacts with mitochondrion and ER. IMM, inner mitochondrial membrane; OMM, outer mitochondrial membrane.
Similar articles
- Universal phase behaviors of intracellular lipid droplets.
Shimobayashi SF, Ohsaki Y. Shimobayashi SF, et al. Proc Natl Acad Sci U S A. 2019 Dec 17;116(51):25440-25445. doi: 10.1073/pnas.1916248116. Epub 2019 Nov 26. Proc Natl Acad Sci U S A. 2019. PMID: 31772016 Free PMC article. - Polarized THG microscopy identifies compositionally different lipid droplets in mammalian cells.
Bautista G, Pfisterer SG, Huttunen MJ, Ranjan S, Kanerva K, Ikonen E, Kauranen M. Bautista G, et al. Biophys J. 2014 Nov 18;107(10):2230-6. doi: 10.1016/j.bpj.2014.10.009. Biophys J. 2014. PMID: 25418291 Free PMC article. - Triglyceride lipolysis triggers liquid crystalline phases in lipid droplets and alters the LD proteome.
Rogers S, Gui L, Kovalenko A, Zoni V, Carpentier M, Ramji K, Ben Mbarek K, Bacle A, Fuchs P, Campomanes P, Reetz E, Speer NO, Reynolds E, Thiam AR, Vanni S, Nicastro D, Henne WM. Rogers S, et al. J Cell Biol. 2022 Nov 7;221(11):e202205053. doi: 10.1083/jcb.202205053. Epub 2022 Sep 16. J Cell Biol. 2022. PMID: 36112368 Free PMC article. - Lipid Droplets in Health and Disease.
Onal G, Kutlu O, Gozuacik D, Dokmeci Emre S. Onal G, et al. Lipids Health Dis. 2017 Jun 29;16(1):128. doi: 10.1186/s12944-017-0521-7. Lipids Health Dis. 2017. PMID: 28662670 Free PMC article. Review. - Plant Lipid Droplets and Their Associated Proteins: Potential for Rapid Advances.
Huang AHC. Huang AHC. Plant Physiol. 2018 Mar;176(3):1894-1918. doi: 10.1104/pp.17.01677. Epub 2017 Dec 21. Plant Physiol. 2018. PMID: 29269574 Free PMC article. Review.
Cited by
- Macrophages release IL11-containing filopodial tip vesicles and contribute to renal interstitial inflammation.
Zhu X, Zhao Y, Liu Y, Shi W, Yang J, Liu Z, Zhang X. Zhu X, et al. Cell Commun Signal. 2023 Oct 18;21(1):293. doi: 10.1186/s12964-023-01327-6. Cell Commun Signal. 2023. PMID: 37853428 Free PMC article. - Cholesterol esters form supercooled lipid droplets whose nucleation is facilitated by triacylglycerols.
Dumesnil C, Vanharanta L, Prasanna X, Omrane M, Carpentier M, Bhapkar A, Enkavi G, Salo VT, Vattulainen I, Ikonen E, Thiam AR. Dumesnil C, et al. Nat Commun. 2023 Feb 17;14(1):915. doi: 10.1038/s41467-023-36375-6. Nat Commun. 2023. PMID: 36807572 Free PMC article. - Seipin-still a mysterious protein?
Salo VT. Salo VT. Front Cell Dev Biol. 2023 Feb 3;11:1112954. doi: 10.3389/fcell.2023.1112954. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36819093 Free PMC article. Review. - Computational Studies of Lipid Droplets.
Kim S, Swanson JMJ, Voth GA. Kim S, et al. J Phys Chem B. 2022 Mar 24;126(11):2145-2154. doi: 10.1021/acs.jpcb.2c00292. Epub 2022 Mar 9. J Phys Chem B. 2022. PMID: 35263109 Free PMC article. Review. - Practical Approaches for Cryo-FIB Milling and Applications for Cellular Cryo-Electron Tomography.
Lam V, Villa E. Lam V, et al. Methods Mol Biol. 2021;2215:49-82. doi: 10.1007/978-1-0716-0966-8_3. Methods Mol Biol. 2021. PMID: 33367999 Free PMC article.
References
- Schuldiner M., Bohnert M., A different kind of love—Lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1188–1196 (2017). - PubMed
- Barbosa A. D., Savage D. B., Siniossoglou S., Lipid droplet-organelle interactions: Emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 35, 91–97 (2015). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials