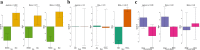Novel age-associated DNA methylation changes and epigenetic age acceleration in middle-aged African Americans and whites - PubMed (original) (raw)
Comparative Study
Novel age-associated DNA methylation changes and epigenetic age acceleration in middle-aged African Americans and whites
Salman M Tajuddin et al. Clin Epigenetics. 2019.
Abstract
Background: African Americans (AAs) experience premature chronic health outcomes and longevity disparities consistent with an accelerated aging phenotype. DNA methylation (DNAm) levels at specific CpG positions are hallmarks of aging evidenced by the presence of age-associated differentially methylated CpG positions (aDMPs) that are the basis for the epigenetic clock for measuring biological age acceleration. Since DNAm has not been widely studied among non-European populations, we examined the association between DNAm and chronological age in AAs and whites, and the association between race, poverty, sex, and epigenetic age acceleration.
Results: We measured genome-wide DNA methylation (866,836 CpGs) using the Illumina MethylationEPIC BeadChip in blood DNA extracted from 487 middle-aged AA (N = 244) and white (N = 243), men (N = 248), and women (N = 239). The mean (sd) age was 48.4 (8.8) in AA and 49.0 (8.7) in whites (p = 0.48). We identified 4930 significantly associated aDMPs in AAs and 469 in whites. Of these, 75.6% and 53.1% were novel, largely driven by the increased number of measured CpGs in the EPIC array, in AA and whites, respectively. AAs had more age-associated DNAm changes than whites in genes implicated in age-related diseases and cellular pathways involved in growth and development. We assessed three epigenetic age acceleration measures (universal, intrinsic, and extrinsic). AAs had a significantly slower extrinsic aging compared to whites. Furthermore, compared to AA women, both AA and white men had faster aging in the universal age acceleration measure (+ 2.04 and + 1.24 years, respectively, p < 0.05).
Conclusions: AAs have more wide-spread methylation changes than whites. Race and sex interact to underlie biological age acceleration suggesting altered DNA methylation patterns may be important in age-associated health disparities.
Keywords: African Americans; Aging; Biological age; DNA methylation; Epigenetic clock; Epigenetics; Epigenome-wide association study; European ancestry; Health disparities; Race.
Conflict of interest statement
MAN’s participation in this study is supported by a consulting contract between Data Tecnica International LLC and the National Institute on Aging (NIA), National Institutes of Health (NIH), Bethesda, Maryland. MAN also consults for Illumina Inc., the Michael J. Fox Foundation, and University of California Healthcare. All other authors declare that they have no competing interests.
Figures
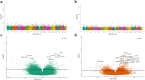
Fig. 1
Distribution of age-associated differentially methylated CpG positions (aDMPs) with their effect size in beta values and significance p value in the African American (AA) and white participants of the HANDLS study: a Manhattan plot in AAs, b Manhattan plot in whites, c volcano plot in AAs, and d volcano plot in whites
Fig. 2
Venn diagrams of significantly age-associated differentially methylated CpG positions (aDMPs) in African Americans (AAs) and whites: a overlap of aDMPs between AAs and whites, b overlap of aDMPs that were hypermethylated with age, and c overlap of aDMPs that were hypomethylated with age
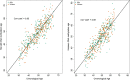
Fig. 3
Correlation between DNA methylation-predicted age based on the Horvath and the Hannum clocks, and chronological age in the HANDLS study. Abbreviation: AAs: African Americans
Fig. 4
Associations between epigenetic age acceleration measures and sex, race, and poverty status. a Sex, b race, and c poverty status. Abbreviations: AAs: African Americans; AgeAccel: universal age acceleration measures; IEAA: intrinsic epigenetic age acceleration; and EEAA: extrinsic epigenetic age acceleration
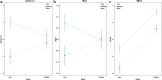
Fig. 5
Interaction plots of the association between sex, race, and three measures of epigenetic age acceleration. a AgeAccel, b IEAA, and c EEAA. Abbreviations: AAs: African Americans; AgeAccel: universal age acceleration measures; IEAA: intrinsic epigenetic age acceleration; and EEAA: extrinsic epigenetic age acceleration
Similar articles
- Race-specific alterations in DNA methylation among middle-aged African Americans and Whites with metabolic syndrome.
Chitrala KN, Hernandez DG, Nalls MA, Mode NA, Zonderman AB, Ezike N, Evans MK. Chitrala KN, et al. Epigenetics. 2020 May;15(5):462-482. doi: 10.1080/15592294.2019.1695340. Epub 2019 Dec 4. Epigenetics. 2020. PMID: 31739726 Free PMC article. - Racial differences in genome-wide methylation profiling and gene expression in breast tissues from healthy women.
Song MA, Brasky TM, Marian C, Weng DY, Taslim C, Dumitrescu RG, Llanos AA, Freudenheim JL, Shields PG. Song MA, et al. Epigenetics. 2015;10(12):1177-87. doi: 10.1080/15592294.2015.1121362. Epigenetics. 2015. PMID: 26680018 Free PMC article. - Systematic evaluation of DNA methylation age estimation with common preprocessing methods and the Infinium MethylationEPIC BeadChip array.
McEwen LM, Jones MJ, Lin DTS, Edgar RD, Husquin LT, MacIsaac JL, Ramadori KE, Morin AM, Rider CF, Carlsten C, Quintana-Murci L, Horvath S, Kobor MS. McEwen LM, et al. Clin Epigenetics. 2018 Oct 16;10(1):123. doi: 10.1186/s13148-018-0556-2. Clin Epigenetics. 2018. PMID: 30326963 Free PMC article. - DNA Methylation as a Biomarker of Aging in Epidemiologic Studies.
Lim U, Song MA. Lim U, et al. Methods Mol Biol. 2018;1856:219-231. doi: 10.1007/978-1-4939-8751-1_12. Methods Mol Biol. 2018. PMID: 30178254 Review. - DNA methylation-based age clocks: From age prediction to age reversion.
Noroozi R, Ghafouri-Fard S, Pisarek A, Rudnicka J, Spólnicka M, Branicki W, Taheri M, Pośpiech E. Noroozi R, et al. Ageing Res Rev. 2021 Jul;68:101314. doi: 10.1016/j.arr.2021.101314. Epub 2021 Mar 5. Ageing Res Rev. 2021. PMID: 33684551 Review.
Cited by
- A cautionary note on altered pace of aging in the COVID-19 era.
Attia MH. Attia MH. Forensic Sci Int Genet. 2022 Jul;59:102724. doi: 10.1016/j.fsigen.2022.102724. Epub 2022 May 17. Forensic Sci Int Genet. 2022. PMID: 35598567 Free PMC article. - Sex-specific transcriptome differences in a middle-aged frailty cohort.
Pacheco NL, Noren Hooten N, Zhang Y, Prince CS, Mode NA, Ezike N, Becker KG, Zonderman AB, Evans MK. Pacheco NL, et al. BMC Geriatr. 2022 Aug 9;22(1):651. doi: 10.1186/s12877-022-03326-7. BMC Geriatr. 2022. PMID: 35945487 Free PMC article. - Epigenetic Aging Biomarkers Associated With Cognitive Impairment in Older African American Adults With Human Immunodeficiency Virus (HIV).
Shiau S, Arpadi SM, Shen Y, Cantos A, Ramon CV, Shah J, Jang G, Manly JJ, Brickman AM, Baccarelli AA, Yin MT. Shiau S, et al. Clin Infect Dis. 2021 Dec 6;73(11):1982-1991. doi: 10.1093/cid/ciab563. Clin Infect Dis. 2021. PMID: 34143869 Free PMC article. - The Socioeconomic Gradient in Epigenetic Ageing Clocks: Evidence from the Multi-Ethnic Study of Atherosclerosis and the Health and Retirement Study.
Schmitz LL, Zhao W, Ratliff SM, Goodwin J, Miao J, Lu Q, Guo X, Taylor KD, Ding J, Liu Y, Levine M, Smith JA. Schmitz LL, et al. Epigenetics. 2022 Jun;17(6):589-611. doi: 10.1080/15592294.2021.1939479. Epub 2021 Jul 6. Epigenetics. 2022. PMID: 34227900 Free PMC article. - Epigenetic aging differentially impacts breast cancer risk by self-reported race.
Wu Y, Miller ME, Gilmore HL, Thompson CL, Schumacher FR. Wu Y, et al. PLoS One. 2024 Oct 24;19(10):e0308174. doi: 10.1371/journal.pone.0308174. eCollection 2024. PLoS One. 2024. PMID: 39446903 Free PMC article.
References
- Tajuddin SM, Amaral AF, Fernandez AF, Rodriguez-Rodero S, Rodriguez RM, Moore LE, Tardon A, Carrato A, Garcia-Closas M, Silverman DT, et al. Genetic and non-genetic predictors of LINE-1 methylation in leukocyte DNA. Environ Health Perspect. 2013;121:650–656. doi: 10.1289/ehp.1206068. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical