Precise Prediction of Calpain Cleavage Sites and Their Aberrance Caused by Mutations in Cancer - PubMed (original) (raw)
Precise Prediction of Calpain Cleavage Sites and Their Aberrance Caused by Mutations in Cancer
Ze-Xian Liu et al. Front Genet. 2019.
Abstract
As a widespread post-translational modification of proteins, calpain-mediated cleavage regulates a broad range of cellular processes, including proliferation, differentiation, cytoskeletal reorganization, and apoptosis. The identification of proteins that undergo calpain cleavage in a site-specific manner is the necessary foundation for understanding the exact molecular mechanisms and regulatory roles of calpain-mediated cleavage. In contrast with time-consuming and labor-intensive experimental methods, computational approaches for detecting calpain cleavage sites have attracted wide attention due to their efficiency and convenience. In this study, we established a novel computational tool named DeepCalpain (http://deepcalpain.cancerbio.info/) for predicting the potential calpain cleavage sites by adopting deep neural network and the particle swarm optimization algorithm. Through critical evaluation and comparison, DeepCalpain exhibited superior performance against other existing tools. Meanwhile, we found that protein interactions could enrich the calpain-substrate regulatory relationship. Since calpain-mediated cleavage was critical for cancer development and progression, we comprehensively analyzed the calpain cleavage associated mutations across 11 cancers with the help of DeepCalpain, which demonstrated that the calpain-mediated cleavage events were affected by mutations and heavily implicated in the regulation of cancer cells. These prediction and analysis results might provide helpful information to reveal the regulatory mechanism of calpain cleavage in biological pathways and different cancer types, which might open new avenues for the diagnosis and treatment of cancers.
Keywords: calpain; cancer mutation; cleavage site; deep learning; prediction.
Figures
Figure 1
Overall methodology. Highlighted are experimentally identified calpain cleavage sites extracted from PubMed by text mining; multi-network deep-learning and PSO algorithm; and the integrative analysis of the connections between calpain-mediated cleavage and cancers.
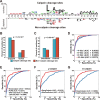
Figure 2
(A) The preferences for the amino acids around the calpain cleavage sites and non-calpain cleavage sites. (B–C) Comparison of the surface accessibility (B) and disorder information between calpain cleavage sites and non-calpain cleavage sites. (D) The 4-, 6-, 8-, and 10-fold cross-validations results for calpain. (E–G) Comparison of the models of calpain (E), µ-calpain (F), and m-calpain (G) with the existing tools, and the dots were the Sn and Sp values adopted from the related literatures.
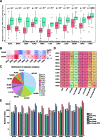
Figure 3
Connections between calpain cleavage sites and genetic variants. (A) Mutations preferentially occur at the regions around calpain cleavage sites. (B) Calpains were differential expressed across cancers. (C) Summary of the distribution of missense variations across cancers. (D) Summary of proteins cleavage aberrant triggered by mutations. (E) Summary of mutation site types across cancers.
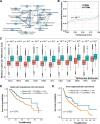
Figure 4
Systematic analysis of the impact of calpain cleavage–related mutation sites and proteins. (A) CCRM proteins were significantly enriched in cancer-related pathways. (B) CCRM sites were more conserved than other mutations. (C) CCRM sites showed a preference to be enriched in known functional domain regions. (D–E) Patients with at least six CCRM sites had significantly worse clinical prognostic in HNSC (D) and LIHC (E).
Similar articles
- GPS-CCD: a novel computational program for the prediction of calpain cleavage sites.
Liu Z, Cao J, Gao X, Ma Q, Ren J, Xue Y. Liu Z, et al. PLoS One. 2011 Apr 20;6(4):e19001. doi: 10.1371/journal.pone.0019001. PLoS One. 2011. PMID: 21533053 Free PMC article. - Deep learning based prediction of reversible HAT/HDAC-specific lysine acetylation.
Yu K, Zhang Q, Liu Z, Du Y, Gao X, Zhao Q, Cheng H, Li X, Liu ZX. Yu K, et al. Brief Bioinform. 2020 Sep 25;21(5):1798-1805. doi: 10.1093/bib/bbz107. Brief Bioinform. 2020. PMID: 32978618 - Deep learning based prediction of species-specific protein S-glutathionylation sites.
Li S, Yu K, Wang D, Zhang Q, Liu ZX, Zhao L, Cheng H. Li S, et al. Biochim Biophys Acta Proteins Proteom. 2020 Jul;1868(7):140422. doi: 10.1016/j.bbapap.2020.140422. Epub 2020 Mar 29. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32234550 - Twenty years of bioinformatics research for protease-specific substrate and cleavage site prediction: a comprehensive revisit and benchmarking of existing methods.
Li F, Wang Y, Li C, Marquez-Lago TT, Leier A, Rawlings ND, Haffari G, Revote J, Akutsu T, Chou KC, Purcell AW, Pike RN, Webb GI, Ian Smith A, Lithgow T, Daly RJ, Whisstock JC, Song J. Li F, et al. Brief Bioinform. 2019 Nov 27;20(6):2150-2166. doi: 10.1093/bib/bby077. Brief Bioinform. 2019. PMID: 30184176 Free PMC article. Review. - Cutting to the chase: calpain proteases in cell motility.
Glading A, Lauffenburger DA, Wells A. Glading A, et al. Trends Cell Biol. 2002 Jan;12(1):46-54. doi: 10.1016/s0962-8924(01)02179-1. Trends Cell Biol. 2002. PMID: 11854009 Review.
Cited by
- Isolation of Platelet-Derived Exosomes from Human Platelet-Rich Plasma: Biochemical and Morphological Characterization.
Saumell-Esnaola M, Delgado D, García Del Caño G, Beitia M, Sallés J, González-Burguera I, Sánchez P, López de Jesús M, Barrondo S, Sánchez M. Saumell-Esnaola M, et al. Int J Mol Sci. 2022 Mar 5;23(5):2861. doi: 10.3390/ijms23052861. Int J Mol Sci. 2022. PMID: 35270001 Free PMC article. - Calpain Regulation and Dysregulation-Its Effects on the Intercalated Disk.
Yoder MW, Wright NT, Borzok MA. Yoder MW, et al. Int J Mol Sci. 2023 Jul 21;24(14):11726. doi: 10.3390/ijms241411726. Int J Mol Sci. 2023. PMID: 37511485 Free PMC article. Review. - Recent Advances Regarding the Molecular Mechanisms of Triterpenic Acids: A Review (Part I).
Mioc M, Milan A, Malița D, Mioc A, Prodea A, Racoviceanu R, Ghiulai R, Cristea A, Căruntu F, Șoica C. Mioc M, et al. Int J Mol Sci. 2022 Jul 13;23(14):7740. doi: 10.3390/ijms23147740. Int J Mol Sci. 2022. PMID: 35887090 Free PMC article. Review. - Effects of truncations in the N- and C-terminal domains of filensin on filament formation with phakinin in cell-free conditions and cultured cells.
Tashiro M, Nakamura A, Kuratani Y, Takada M, Iwamoto S, Oka M, Ando S. Tashiro M, et al. FEBS Open Bio. 2023 Nov;13(11):1990-2004. doi: 10.1002/2211-5463.13700. Epub 2023 Aug 30. FEBS Open Bio. 2023. PMID: 37615966 Free PMC article. - Calpain-1 weakens the nuclear envelope and promotes the release of neutrophil extracellular traps.
Singh J, Zlatar L, Muñoz-Becerra M, Lochnit G, Herrmann I, Pfister F, Janko C, Knopf J, Leppkes M, Schoen J, Muñoz LE, Schett G, Herrmann M, Schauer C, Mahajan A. Singh J, et al. Cell Commun Signal. 2024 Sep 9;22(1):435. doi: 10.1186/s12964-024-01785-6. Cell Commun Signal. 2024. PMID: 39252008 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials