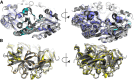Structure of a Synthetic β-Carboxysome Shell - PubMed (original) (raw)
Structure of a Synthetic _β_-Carboxysome Shell
Markus Sutter et al. Plant Physiol. 2019 Nov.
Abstract
Carboxysomes are capsid-like, CO2-fixing organelles that are present in all cyanobacteria and some chemoautotrophs and that substantially contribute to global primary production. They are composed of a selectively permeable protein shell that encapsulates Rubisco, the principal CO2-fixing enzyme, and carbonic anhydrase. As the centerpiece of the carbon-concentrating mechanism, by packaging enzymes that collectively enhance catalysis, the carboxysome shell enables the generation of a locally elevated concentration of substrate CO2 and the prevention of CO2 escape. A functional carboxysome consisting of an intact shell and cargo is essential for cyanobacterial growth under ambient CO2 concentrations. Using cryo-electron microscopy, we have determined the structure of a recombinantly produced simplified β-carboxysome shell. The structure reveals the sidedness and the specific interactions between the carboxysome shell proteins. The model provides insight into the structural basis of selective permeability of the carboxysome shell and can be used to design modifications to investigate the mechanisms of cargo encapsulation and other physiochemical properties such as permeability. Notably, the permeability properties are of great interest for modeling and evaluating this carbon-concentrating mechanism in metabolic engineering. Moreover, we find striking similarity between the carboxysome shell and the structurally characterized, evolutionarily distant metabolosome shell, implying universal architectural principles for bacterial microcompartment shells.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures
Figure 1.
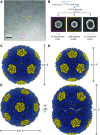
Cryo-EM reconstruction of synthetic shells. A, Example cryo-EM micrograph used in reconstruction with example particles of different shell types boxed (T = 4 in magenta, T = 3 in yellow, and T = 4, Q = 6 prolate in cyan). All particles were used for reconstruction, but only selected ones are highlighted for clarity. B, Single-particle analysis preprocessing workflow with example 2D class averages for the different shell types. Particle counts and percentage of input particles for each class are indicated. C to E, 3D reconstructions for the T = 4 (C), T = 3 (D), and prolate (E) shell types with dimensions indicated.
Figure 2.
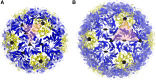
Geometrical description of the regular icosahedral shell types. Closeup of the cartoon model for each shell type is shown with the asymmetric unit indicated by red triangles. Subunits corresponding to the T = 3 and T = 4 lattices are indicated by solid lines for the icosahedral shells in A and B, respectively.
Figure 3.
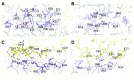
Protein subunit interfaces. A and B, View onto the CcmK-CcmK interface from the outside (A) and from the inside (B). C and D, View of the CcmK-CcmL interface from the inside (C) and outside (D). Interface residues are shown as sticks, CcmK in shades of blue and CcmL in yellow. Important residues are labeled, and dashed lines indicate specific interactions.
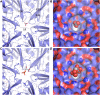
Figure 4.
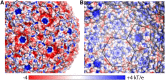
Electrostatics of the T = 4 shell. A, Surface as seen from the outside colored by electrostatic potential (red, negative; blue, positive), with outlines of CcmK and CcmL indicated. B, Same as A, but for the inside view. The scale bar below indicates the range from −4 to +4 kT/e.
Figure 5.
Structural alignment of Halothece sp. PCC 7418 BMC-H and BMC-P from the shell with corresponding isolated structures of other β-carboxysomes. A, Alignment of Halothece sp. PCC 7418 CcmK1 and CcmK2 (blue), with available crystal structures of CcmK1 and CcmK2 (Protein Data Bank identifiers 2A1B, 3BN4, 3CIM, 3SSQ, 3SSS, and 4OX7) in different shades of gray. Views of the aligned chain in cyan (left) and opposite side (right) are shown. B, Alignment of Halothece sp. PCC 7418 CcmL (yellow), with homologous structures (Protein Data Bank identifiers 2QW7, 4JVZ, 4JW0, and 4N8X) in different shades of gray.
Figure 6.
A, Closeup of the CcmK pore of the Halothece sp. PCC 7418 shell with a bicarbonate modeled in the center. K36 lines the concave side of the pore, and hydrogen bonds to the E35 side chain and the backbone oxygen of K36 of the adjacent chain. B, Surface representation of A. C and D, Same as A and B, but with PGA.
Similar articles
- Rubisco packaging and stoichiometric composition of the native β-carboxysome in Synechococcus elongatus PCC7942.
Sun Y, Sheng Y, Ni T, Ge X, Sarsby J, Brownridge PJ, Li K, Hardenbrook N, Dykes GF, Rockliffe N, Eyers CE, Zhang P, Liu LN. Sun Y, et al. Plant Physiol. 2024 Dec 24;197(1):kiae665. doi: 10.1093/plphys/kiae665. Plant Physiol. 2024. PMID: 39680612 Free PMC article. - Uncovering the roles of the scaffolding protein CsoS2 in mediating the assembly and shape of the α-carboxysome shell.
Li T, Chen T, Chang P, Ge X, Chriscoli V, Dykes GF, Wang Q, Liu L-N. Li T, et al. mBio. 2024 Oct 16;15(10):e0135824. doi: 10.1128/mbio.01358-24. Epub 2024 Aug 29. mBio. 2024. PMID: 39207096 Free PMC article. - Identification of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome.
Blikstad C, Dugan EJ, Laughlin TG, Turnšek JB, Liu MD, Shoemaker SR, Vogiatzi N, Remis JP, Savage DF. Blikstad C, et al. Proc Natl Acad Sci U S A. 2023 Oct 24;120(43):e2308600120. doi: 10.1073/pnas.2308600120. Epub 2023 Oct 20. Proc Natl Acad Sci U S A. 2023. PMID: 37862384 Free PMC article. - Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells.
Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Yeates TO, et al. Biochem Soc Trans. 2007 Jun;35(Pt 3):508-11. doi: 10.1042/BST0350508. Biochem Soc Trans. 2007. PMID: 17511640 Review. - Role of carboxysomes in cyanobacterial CO2 assimilation: CO2 concentrating mechanisms and metabolon implications.
Huffine CA, Zhao R, Tang YJ, Cameron JC. Huffine CA, et al. Environ Microbiol. 2023 Feb;25(2):219-228. doi: 10.1111/1462-2920.16283. Epub 2022 Nov 22. Environ Microbiol. 2023. PMID: 36367380 Review.
Cited by
- Recent structural insights into bacterial microcompartment shells.
Ochoa JM, Yeates TO. Ochoa JM, et al. Curr Opin Microbiol. 2021 Aug;62:51-60. doi: 10.1016/j.mib.2021.04.007. Epub 2021 May 28. Curr Opin Microbiol. 2021. PMID: 34058518 Free PMC article. Review. - A catalog of the diversity and ubiquity of bacterial microcompartments.
Sutter M, Melnicki MR, Schulz F, Woyke T, Kerfeld CA. Sutter M, et al. Nat Commun. 2021 Jun 21;12(1):3809. doi: 10.1038/s41467-021-24126-4. Nat Commun. 2021. PMID: 34155212 Free PMC article. - Towards using bacterial microcompartments as a platform for spatial metabolic engineering in the industrially important and metabolically versatile Zymomonas mobilis.
Doron L, Raval D, Kerfeld CA. Doron L, et al. Front Bioeng Biotechnol. 2024 Jan 26;12:1344260. doi: 10.3389/fbioe.2024.1344260. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38344288 Free PMC article. - A robust synthetic biology toolkit to advance carboxysome study and redesign.
Trettel DS, Hoang Y, Vecchiarelli AG, Gonzalez-Esquer CR. Trettel DS, et al. bioRxiv [Preprint]. 2024 Oct 8:2024.10.08.617227. doi: 10.1101/2024.10.08.617227. bioRxiv. 2024. PMID: 39416180 Free PMC article. Updated. Preprint. - Molecular interactions of the chaperone CcmS and carboxysome shell protein CcmK1 that mediate β-carboxysome assembly.
Cheng J, Li CY, Meng M, Li JX, Liu SJ, Cao HY, Wang N, Zhang YZ, Liu LN. Cheng J, et al. Plant Physiol. 2024 Nov 4;196(3):1778-1787. doi: 10.1093/plphys/kiae438. Plant Physiol. 2024. PMID: 39172695 Free PMC article.
References
- Biyani N, Righetto RD, McLeod R, Caujolle-Bert D, Castano-Diez D, Goldie KN, Stahlberg H (2017) Focus: The interface between data collection and data processing in cryo-EM. J Struct Biol 198: 124–133 - PubMed
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell 155: 1131–1140 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials