Geographical and Ethnic Differences Influence Culturable Commensal Yeast Diversity on Healthy Skin - PubMed (original) (raw)
Geographical and Ethnic Differences Influence Culturable Commensal Yeast Diversity on Healthy Skin
Cheryl Leong et al. Front Microbiol. 2019.
Abstract
Commensal fungi such as Malassezia, Candida, and Rhodotorula are common on healthy skin but are also associated with opportunistic invasive and superficial infections. Skin microbial community characterization has been extensively performed worldwide, with a focus on the 16S bacterial community. These studies have focused on geographically distinct or targeted cohorts with variable reported species distributions of commensal yeast species. To determine the effects of extrinsic environmental factors such as geography, climate, and ethnicity on detected healthy skin commensal yeast diversity, we compared cohorts from Singapore and Zürich, Switzerland, representative of two geographically and climatically distinct regions comprising multi-ethnic (Chinese, Malay, Indian, Caucasian) and predominantly white Caucasian cohorts, respectively, using identical skin sampling and culture methods. We chose to use a culture-based approach as cultures isolated from patients are still required for studies of pathogenicity and antifungal susceptibility. Detection of yeast species by culture-dependent and independent sequencing-based methods suggest healthy skin diversity reflects a species distribution representative of the geography, climate and ethnic background of their local populations. Culture success and species diversity was also found to be dependent on climate, with warm tropical climates favoring high positive culture rates and greater species diversity. Multilocus sequence typing data suggests some strains are geographically distinct and may be used to segregate potential disease-causing commensals. For accurate collection and characterization of skin microbial communities, it remains recommended to employ a combination of culture-dependent and sequence-based culture-independent methods. Characterization of healthy mycobiomes in geographically distinct local populations will be useful in defining the role of commensal fungi in health and disease.
Keywords: Malassezia; commensal yeast; ethnicity; fungi; geography; skin.
Figures
FIGURE 1
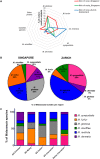
(A) Radar plot of percentage of subjects that were culture positive for each species (_y_-axis) as sampled from the respective cohorts and body sites, (B) pie charts showing percentage of each Malassezia species isolated from the skin of the nose calculated as a percentage of total Malassezia isolates from each respective cohort, and (C) Malassezia species isolated from the skin of nose of Singapore (n = 37, 10 Chinese, 7 Malay, 10 Indian, and 10 Caucasian, equal gender) and Swiss (n = 11) cohort subjects, represented as a percentage of the total Malassezia species isolated from each ethnic group.
FIGURE 2
(A) Multiple Correspondence Analysis (MCA) plot showing the clustering of 48 individuals form Singapore and Zürich, Switzerland based on geography, gender, race, and Malassezia species cultured from the skin of the nose, (B) factor maps of individual clustering by gender (M – Male, F – Female), geography (SG – Singapore, ZH – Zürich), and race (Ch – Chinese, Ma – Malay, Ind – Indian, Ca – Caucasian) as defined by a 95% confidence ellipse, and (C) clustering dendrogram showing the clustering of individuals by hierarchical clustering. Numbering of individuals correspond to the legend in (A). Circles indicate individuals that do not cluster according to their expected geography or race.
FIGURE 3
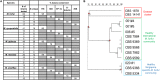
(A) Table comparison of commensal yeast species detection by culture-dependent and culture independent methods in four subjects and (B) PCR detection using species-specific primers of gDNA isolated from the parallel side of the skin of the nose from which cultures were swab-derived. gDNA isolated from reference strain cultures representative of common isolates of Malassezia (CBS 9369, CBS 7222, CBS 7966, CBS 7877, CBS 7956), Rhodotorula (ATCC 9449), and Candida (ATCC 22019) were used as positive controls. A 100 bp ladder (GeneRulerTM) was run on the leftmost column of each gel.
FIGURE 4
(A) Percentage conserved bases of five commonly isolated species of Malassezia across five loci (Internal transcribed spacer region – ITS, Intergenic spacer region (IGS), Elongation factor – 1α (EF-1α), ß-tubulin and the 26S ribosomal region) as tested across sequences derived from isolates collected and NCBI databases and (B) unrooted dendrogram derived from 13 unique ITS sequences of M. furfur identify healthy and disease clusters which are representative of strains isolated in Singapore and internationally. Sequences not represented by any known reference database ID are indicated by their subject/strain codes.
Similar articles
- Comparison of Three Skin Sampling Methods and Two Media for Culturing Malassezia Yeast.
Abdillah A, Khelaifia S, Raoult D, Bittar F, Ranque S. Abdillah A, et al. J Fungi (Basel). 2020 Dec 9;6(4):350. doi: 10.3390/jof6040350. J Fungi (Basel). 2020. PMID: 33316902 Free PMC article. - Malassezia species in healthy skin and in dermatological conditions.
Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. Prohic A, et al. Int J Dermatol. 2016 May;55(5):494-504. doi: 10.1111/ijd.13116. Epub 2015 Dec 29. Int J Dermatol. 2016. PMID: 26710919 Review. - Frequency, body distribution, and population size of Malassezia species in healthy dogs and in dogs with localized cutaneous lesions.
Cafarchia C, Gallo S, Romito D, Capelli G, Chermette R, Guillot J, Otranto D. Cafarchia C, et al. J Vet Diagn Invest. 2005 Jul;17(4):316-22. doi: 10.1177/104063870501700403. J Vet Diagn Invest. 2005. PMID: 16130988 - Candida and candidaemia. Susceptibility and epidemiology.
Arendrup MC. Arendrup MC. Dan Med J. 2013 Nov;60(11):B4698. Dan Med J. 2013. PMID: 24192246 Review.
Cited by
- The human pathobiont Malassezia furfur secreted protease Mfsap1 regulates cell dispersal and exacerbates skin inflammation.
Goh JPZ, Ruchti F, Poh SE, Koh WLC, Tan KY, Lim YT, Thng STG, Sobota RM, Hoon SS, Liu C, O'Donoghue AJ, LeibundGut-Landmann S, Oon HH, Li H, Dawson TL Jr. Goh JPZ, et al. Proc Natl Acad Sci U S A. 2022 Dec 6;119(49):e2212533119. doi: 10.1073/pnas.2212533119. Epub 2022 Nov 29. Proc Natl Acad Sci U S A. 2022. PMID: 36442106 Free PMC article. - Identification of Malassezia furfur Secreted Aspartyl Protease 1 (MfSAP1) and Its Role in Extracellular Matrix Degradation.
Poh SE, Goh JPZ, Fan C, Chua W, Gan SQ, Lim PLK, Sharma B, Leavesley DI, Dawson TL Jr, Li H. Poh SE, et al. Front Cell Infect Microbiol. 2020 Apr 9;10:148. doi: 10.3389/fcimb.2020.00148. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32328468 Free PMC article. - Truncal acne following JAK inhibitor use in vitiligo with rare opportunistic fungal infections: Two case reports.
Doh JY, Rintarhat P, Jung WH, Kim HS. Doh JY, et al. JAAD Case Rep. 2023 May 23;37:123-127. doi: 10.1016/j.jdcr.2023.05.018. eCollection 2023 Jul. JAAD Case Rep. 2023. PMID: 37405176 Free PMC article. No abstract available. - Malassezia: Zoonotic Implications, Parallels and Differences in Colonization and Disease in Humans and Animals.
Hobi S, Cafarchia C, Romano V, Barrs VR. Hobi S, et al. J Fungi (Basel). 2022 Jul 4;8(7):708. doi: 10.3390/jof8070708. J Fungi (Basel). 2022. PMID: 35887463 Free PMC article. Review. - State of the Art in the Culture of the Human Microbiota: New Interests and Strategies.
Tidjani Alou M, Naud S, Khelaifia S, Bonnet M, Lagier JC, Raoult D. Tidjani Alou M, et al. Clin Microbiol Rev. 2020 Oct 28;34(1):e00129-19. doi: 10.1128/CMR.00129-19. Print 2020 Dec 16. Clin Microbiol Rev. 2020. PMID: 33115723 Free PMC article. Review.
References
- Chang H. J., Miller H. L., Watkins N., Arduino M. J., Ashford D. A., Midgley G., et al. (1998). An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonization of health care workers’ pet dogs. N. Engl. J. Med. 338 706–711. 10.1056/NEJM199803123381102 - DOI - PubMed
LinkOut - more resources
Full Text Sources