Optical control of protein phosphatase function - PubMed (original) (raw)
Optical control of protein phosphatase function
Taylor M Courtney et al. Nat Commun. 2019.
Abstract
Protein phosphatases are involved in embryonic development, metabolic homeostasis, stress response, cell cycle transitions, and many other essential biological mechanisms. Unlike kinases, protein phosphatases remain understudied and less characterized. Traditional genetic and biochemical methods have contributed significantly to our understanding; however, these methodologies lack precise and acute spatiotemporal control. Here, we report the development of a light-activated protein phosphatase, the dual specificity phosphatase 6 (DUSP6 or MKP3). Through genetic code expansion, MKP3 is placed under optical control via two different approaches: (i) incorporation of a caged cysteine into the active site for controlling catalytic activity and (ii) incorporation of a caged lysine into the kinase interaction motif for controlling the protein-protein interaction between the phosphatase and its substrate. Both strategies are expected to be applicable to the engineering of a wide range of light-activated phosphatases. Applying the optogenetically controlled MKP3 in conjunction with live cell reporters, we discover that ERK nuclear translocation is regulated in a graded manner in response to increasing MKP3 activity.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
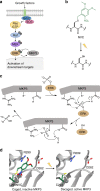
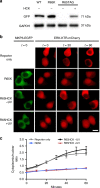
Overview of the MAPK/MKP3 signaling network and application of caged cysteine toward caging the catalytic site of the phosphatase. a Schematic of the Ras/Raf/ERK signaling pathway with the dual-specificity phosphatase MKP3 indicated in grey. Upon activation of the pathway by external stimuli, a cascade of phosphorylation events results in dually-phosphorylated ERK, which translocates to the nucleus where it activates additional downstream targets. In the presence of active MKP3, pERK is rapidly dephosphorylated and remains in the cytoplasm. Phosphates are indicated by grey filled circles. b Irradiation of NVC removes the nitrobenzyl caging group (green) and restores a native cysteine residue. c The mechanism of MKP3-catalyzed ERK dephosphorylation. Dashed lines indicate electrostatic and/or hydrogen-bond interactions. d The crystal structure of MKP3 (PDB: 1MKP) is shown with the three critical active site residues labeled in black. On the left, the catalytic cysteine was replaced with NVC, which sterically blocks the active site and prevents catalysis, until exposed to 365 nm light
Fig. 2
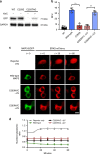
Optical control of the MKP3 catalytic site. a Western blot analysis confirms expression of caged MKP3 only in the presence of NVC (1 mM). b Biochemical phosphatase assays show complete loss of activity for the caged enzyme, until 365 nm irradiation restores the wild-type enzyme. Error bars denote standard deviation from three biological replicates; **p < 0.01 from unpaired two-tailed Student’s _t_-test. **c** Using an ERK2-mCherry reporter, the activity of wild-type, caged (at the catalytic cysteine), and photoactivated MKP3 was investigated in HEK293T cells. Following pathway stimulation, nuclear exclusion of an ERK2-mCherry reporter is observed for all MKP3 variants. Scale bar 10 μm; also see Supplementary Fig. 6. **d** Quantification of nuclear/cytoplasmic ratios shows successful nuclear translocation for the reporter only control; however, none of the MKP3 variants show any significant change. Error bars represent standard error of the mean from nine individual cells combined from three biological replicates. One-way ANOVA tests at the 60-minute time point results in _p_ < 0.0001 for the reporter compared to all MKP3 samples, indicating this a significant difference. Comparing C293NVC –UV to C293NVC + UV results in _p_ > 0.1, indicating no significant difference. Data are provided in the Source Data file
Fig. 3
ERK activity in response to optical activation of the MKP3 catalytic site. a An ERK-KTR-mCherry reporter was utilized to report on the activity of endogenous ERK2 levels in the presence of wild-type, caged, and photoactivated MKP3 variants in HEK293T cells. Wild-type MKP3 prevents EGF-induced triggering of the reporter, while the caged MKP3 is only active following UV exposure (365 nm, 2 min). Scale bar 10 μm; also see Supplementary Fig. 7. b Quantification of the cytoplasmic/nuclear ratio for the MKP3 variants shows successful photoactivation of MKP3. Error bars represent standard error of the mean for nine individual cells combined from three biological replicates. One-way ANOVA tests at the 60-minute time point resulted in p < 0.0005 when comparing C293NVC –UV to C293NVC + UV, and reporter only to WT. No significant difference (_p_ > 0.1) was observed between WT and C293NVC + UV or reporter only and C293NVC –UV. Data are provided in the Source Data file
Fig. 4
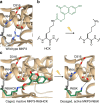
Design strategy for optical control of the phosphatase-kinase interaction motif (KIM). a The crystal structure of ERK2 (tan) bound to the KIM peptide of MKP3 (gray) shows essential electrostatic interactions of R64 and R65 with D316 and D319 (PDB: 2FYS). b Irradiation of HCK removes the coumarin caging group (green) and restores a native lysine residue. c Replacement of R65 with HCK generates steric hindrance and prevents electrostatic interaction until light exposure release K65, which enables the ERK2-MKP3 interaction
Fig. 5
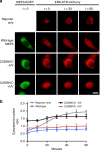
ERK activity in response to optical activation of the MKP3 kinase interaction motif. a Western blot of wild-type, R65K, and caged MKP3 shows full-length expression only in the presence of 0.25 mM HCK. b KIM-caged MKP3 cannot efficiently bind to and dephosphorylate pERK until UV irradiation (365 nm, 2 min) generates an active enzyme. Scale bar 10 μm; also see Supplementary Fig. 8. c) Quantification of cytoplasmic-to-nuclear (C/N) ratios support the micrograph findings. Error bars represent standard error of the mean of nine individual cells from three biological replicates. One-way ANOVA tests at the 60-minute time point resulted in p < 0.0001 when comparing R65HCK –UV to R65HCK + UV, and reporter only to R65K. No significant difference (_p_ > 0.1) was observed between R65K and R65HCK + UV or reporter only and R65HCK –UV. Data are provided in the Source Data file
Fig. 6
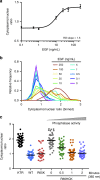
Analysis of ERK activity in response to different doses of growth factor stimulation and optically tuned phosphatase function. a The cytoplasmic/nuclear ratios for the ERK-KTR-mCherry reporter in HEK293T cells were plotted for a range of EGF concentrations and fitted with a Hill function. Based on the calculated Hill slope, a graded response is observed. Error bars represent the standard error of the mean of 85–90 cells per condition pooled from three biological replicates. b Histogram representation of the obtained data more clearly depicts the graded response as seen by a unimodal distribution. c Optical activation of KIM-caged MKP3 shows a graded response for the regulation of pERK translocation upon increased UV irradiation and cell surface stimulation with EGF (100 ng/mL), and thus increased levels of active MKP3. Each dot represents a single cell, black lines represent the mean, n = 40–45. Quantified cells were pooled from three biological replicates. One-way ANOVA tests showed p < 0.0001 comparing 0 min to 0.5 min irradiation, _p_ = 0.0002 for 0.5–1 min irradiation, and _p_ > 0.1 for 1–2 min irradiation, indicating a significant difference for the first two comparisons and no significant difference for the third. Data are provided in the Source Data file
Comment in
- Controlling Phosphate Removal with Light: The Development of Optochemical Tools to Probe Protein Phosphatase Function.
Courtney TM, Deiters A. Courtney TM, et al. SLAS Discov. 2020 Sep;25(8):957-960. doi: 10.1177/2472555220918519. Epub 2020 May 14. SLAS Discov. 2020. PMID: 32406806
Similar articles
- Caspase-3 cleavage of DUSP6/MKP3 at the interdomain region generates active MKP3 fragments that regulate ERK1/2 subcellular localization and function.
Cejudo-Marín R, Tárrega C, Nunes-Xavier CE, Pulido R. Cejudo-Marín R, et al. J Mol Biol. 2012 Jun 29;420(1-2):128-38. doi: 10.1016/j.jmb.2012.04.004. Epub 2012 Apr 11. J Mol Biol. 2012. PMID: 22504224 - Structural and Dynamic Insights into the Mechanism of Allosteric Signal Transmission in ERK2-Mediated MKP3 Activation.
Lu C, Liu X, Zhang CS, Gong H, Wu JW, Wang ZX. Lu C, et al. Biochemistry. 2017 Nov 21;56(46):6165-6175. doi: 10.1021/acs.biochem.7b00827. Epub 2017 Nov 8. Biochemistry. 2017. PMID: 29077400 - Optical control of biological processes by light-switchable proteins.
Fan LZ, Lin MZ. Fan LZ, et al. Wiley Interdiscip Rev Dev Biol. 2015 Sep-Oct;4(5):545-54. doi: 10.1002/wdev.188. Epub 2015 Apr 8. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25858669 Free PMC article. Review. - Synergistic Ensemble of Optogenetic Actuators and Dynamic Indicators in Cell Biology.
Kim J, Heo WD. Kim J, et al. Mol Cells. 2018 Sep 30;41(9):809-817. doi: 10.14348/molcells.2018.0295. Epub 2018 Aug 29. Mol Cells. 2018. PMID: 30157546 Free PMC article. Review.
Cited by
- Applications of genetic code expansion technology in eukaryotes.
Guo QR, Cao YJ. Guo QR, et al. Protein Cell. 2024 May 7;15(5):331-363. doi: 10.1093/procel/pwad051. Protein Cell. 2024. PMID: 37847216 Free PMC article. Review. - Mining oomycete proteomes for phosphatome leads to the identification of specific expanded phosphatases in oomycetes.
Qiu M, Sun Y, Tu S, Li H, Yang X, Zhao H, Yin M, Li Y, Ye W, Wang M, Wang Y. Qiu M, et al. Mol Plant Pathol. 2024 Mar;25(3):e13425. doi: 10.1111/mpp.13425. Mol Plant Pathol. 2024. PMID: 38462784 Free PMC article. - Targeted protein oxidation using a chromophore-modified rapamycin analog.
Courtney TM, Hankinson CP, Horst TJ, Deiters A. Courtney TM, et al. Chem Sci. 2021 Sep 22;12(40):13425-13433. doi: 10.1039/d1sc04464h. eCollection 2021 Oct 20. Chem Sci. 2021. PMID: 34777761 Free PMC article. - The Development and Application of Opto-Chemical Tools in the Zebrafish.
Feng Z, Ducos B, Scerbo P, Aujard I, Jullien L, Bensimon D. Feng Z, et al. Molecules. 2022 Sep 22;27(19):6231. doi: 10.3390/molecules27196231. Molecules. 2022. PMID: 36234767 Free PMC article. Review. - Photochemical modifications for DNA/RNA oligonucleotides.
Tavakoli A, Min JH. Tavakoli A, et al. RSC Adv. 2022 Feb 24;12(11):6484-6507. doi: 10.1039/d1ra05951c. eCollection 2022 Feb 22. RSC Adv. 2022. PMID: 35424630 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous