A four-gene signature-derived risk score for glioblastoma: prospects for prognostic and response predictive analyses - PubMed (original) (raw)
. 2019 Aug;16(3):595-605.
doi: 10.20892/j.issn.2095-3941.2018.0277.
Juan Cai 2, Ye Yuan 1, Yu Shi 1, Hong Wu 1, Qing Liu 1, Yueliang Yao 1, Lu Chen 1, Weiqi Dang 1, Xiang Zhang 1, Jingfang Xiao 1, Kaidi Yang 1, Zhicheng He 1, Xiaohong Yao 1, Yonghong Cui 1, Xia Zhang 1, Xiuwu Bian 1
Affiliations
- PMID: 31565488
- PMCID: PMC6743613
- DOI: 10.20892/j.issn.2095-3941.2018.0277
A four-gene signature-derived risk score for glioblastoma: prospects for prognostic and response predictive analyses
Mianfu Cao et al. Cancer Biol Med. 2019 Aug.
Abstract
Objective: Glioblastoma (GBM) is the most common primary malignant brain tumor regulated by numerous genes, with poor survival outcomes and unsatisfactory response to therapy. Therefore, a robust, multi-gene signature-derived model is required to predict the prognosis and treatment response in GBM.
Methods: Gene expression data of GBM from TCGA and GEO datasets were used to identify differentially expressed genes (DEGs) through DESeq2 or LIMMA methods. The DEGs were then overlapped and used for survival analysis by univariate and multivariate COX regression. Based on the gene signature of multiple survival-associated DEGs, a risk score model was established, and its prognostic and predictive role was estimated through Kaplan-Meier analysis and log-rank test. Gene set enrichment analysis (GSEA) was conducted to explore high-risk score-associated pathways. Western blot was used for protein detection.
Results: Four survival-associated DEGs of GBM were identified: OSMR, HOXC10, SCARA3, and SLC39A10. The four-gene signature-derived risk score was higher in GBM than in normal brain tissues. GBM patients with a high-risk score had poor survival outcomes. The high-risk group treated with temozolomide chemotherapy or radiotherapy survived for a shorter duration than the low-risk group. GSEA showed that the high-risk score was enriched with pathways such as vasculature development and cell adhesion. Western blot confirmed that the proteins of these four genes were differentially expressed in GBM cells.
Conclusions: The four-gene signature-derived risk score functions well in predicting the prognosis and treatment response in GBM and will be useful for guiding therapeutic strategies for GBM patients.
Keywords: Differentially expressed genes; gene set enrichment analysis; glioblastoma prognosis; radiotherapy; temozolomide chemotherapy.
Copyright 2019 Cancer Biology & Medicine.
Figures
1
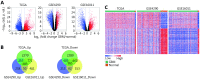
Differentially expressed genes between GBM and normal brain tissues. (A) Volcano plots showing the log2 (fold change) of mRNA in GBM compared to normal brain tissues, and the corresponding–log10 (adjusted P value) in TCGA, GSE4290 and GSE16011 datasets. Genes with adjusted P value below 0.05 and fold change above 2 (below -2) were marked with red (blue) dots. (B) Venn diagrams showing the gene numbers of the upregulated genes (left) and the downregulated genes (right) of GBM in TCGA, GSE4290 and GSE16011 cohorts. (C) Heatmaps of the overlapped genes in TCGA, GSE4290 and GSE16011 datasets. The overlapped genes that are from Figure 1B include 483 upregulated genes and 765 downregulated genes.
2
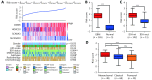
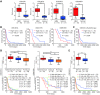
The four-gene signature-derived risk score is high in GBM. (A) The calculation formula and the value of risk score (top), the corresponding expression of four key genes (middle), and the associated clinicopathological parameters (bottom). “β” indicates the regression coefficient derived from multivariate COX stepwise regression in TCGA GBM cohort; “E” represents the expression value of the corresponding gene. The _P_-value indicates the correlation between the risk score and the clinicopathological parameters (Supplementary Table S5). KPS, Karnofsky performance status; IDH, isocitrate dehydrogenase; MGMT, O(6)-methylguanine-DNA methyltransferase; NA, not available. (B-D) Risk scores in GBM and normal brain tissues (B), in IDH-wt GBM and IDH-mut GBM (C), and in mesenchymal GBM and non-mesenchymal (proneural and classical) GBM (D) in TCGA GBM cohort. IDH, isocitrate dehydrogenase; IDH-wt, IDH-wild type; IDH-mut, IDH-mutation. Data are shown as mean ± SEM, *P < 0.05, *** P < 0.001, ns, not significant.
S1
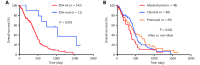
Survival analysis of GBM patients. (A-B) Kaplan-Meier overall survival analysis of GBM patients stratified by IDH status (A), and expression subtypes (B). IDH, isocitrate dehydrogenase; IDH-wt, IDH-wild type; IDH-mut, IDH-mutation. Mes, mesenchymal; Non-Mes, non-mesenchymal, including proneural and classical subtypes.
3
Risk score predicts the prognosis and treatment response in TCGA GBM cohort. (A-C) Kaplan–Meier overall survival analysis among TCGA GBM patients stratified by risk score only (A), combined with IDH status (B), and combined with expression subtypes (C). Mes, mesenchymal; Non-Mes, non-mesenchymal, including proneural and classical subtypes. (D-E) Kaplan–Meier overall survival analysis of TCGA GBM patients with TMZ chemotherapy (D), or radiotherapy (E) according to the risk score. TMZ, temozolomide.
4
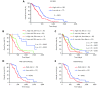
Performance of risk score in predicting the survival and treatment response in independent GEO GBM cohorts. (A) Comparison of risk scores between GBM and normal brain tissues in GSE4290, GSE16011, GSE59612 and GSE90604. (B) Kaplan-Meier overall survival analysis of high-risk and low-risk GBM patients in GSE16011, GSE43378 and GSE83300. (C) Kaplan-Meier overall survival analysis of GBM patients with radiotherapy in GSE16011 according to the risk score. (D) Risk scores in mesenchymal GBM and non-mesenchymal (proneural and classical) GBM (up), and Kaplan–Meier overall survival analysis of GBM patients stratified by risk scores combined with expression subtypes (down). Mes, mesenchymal; Non-Mes, non-mesenchymal, including proneural and classical subtypes. (E) Risk scores in IDH-wt GBM and IDH-mut GBM (up), and Kaplan–Meier overall survival analysis of GBM patients stratified by risk scores combined with IDH status (down). IDH, isocitrate dehydrogenase; IDH-wt, IDH-wild type; IDH-mut, IDH-mutation. Data are shown as mean ± SEM, *P < 0.05, ** P < 0.01, *** P < 0.001, ns, not significant.
5
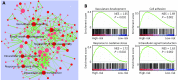
Four-gene signature associated biological pathways in high-risk GBM. (A) Enrichment map showing the pathways enriched in high-risk GBM through GSEA analysis. Nodes represent enriched gene sets with _P_-value below 0.05. Node with deep red color correlates with small _P_-value. Node size corresponds to the number of genes within gene set. Edge thickness corresponds to the number of shared genes between gene sets. (B) Representative enriched pathways in high-risk GBM through GSEA analysis. NES, normalized enrichment score.
6
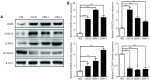
Validation of the differential expression of the four genes in GBM cells. (A and B) Western blot images (A) and the relevant quantification (B) of OSMR, HOXC10, SCARA3 and SLC39A10 in GBM cell line LN229, primary GBM cells (GBM-1 and GBM-2), and normal glial cell line HEB. The relative expression of target proteins is quantified in comparison with β-Actin and normalized to the corresponding expression in HEB cells. Data are shown as mean ± SEM from three independent experiments, *P < 0.05, ** P < 0.01, *** P < 0.001.
Similar articles
- Angiogenesis-Related Gene Signature-Derived Risk Score for Glioblastoma: Prospects for Predicting Prognosis and Immune Heterogeneity in Glioblastoma.
Wang G, Hu JQ, Liu JY, Zhang XM. Wang G, et al. Front Cell Dev Biol. 2022 Mar 18;10:778286. doi: 10.3389/fcell.2022.778286. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35372355 Free PMC article. - Large-Scale Analysis Reveals Gene Signature for Survival Prediction in Primary Glioblastoma.
Prasad B, Tian Y, Li X. Prasad B, et al. Mol Neurobiol. 2020 Dec;57(12):5235-5246. doi: 10.1007/s12035-020-02088-w. Epub 2020 Sep 1. Mol Neurobiol. 2020. PMID: 32875483 Free PMC article. - A Five-lncRNAs Signature-Derived Risk Score Based on TCGA and CGGA for Glioblastoma: Potential Prospects for Treatment Evaluation and Prognostic Prediction.
Niu X, Sun J, Meng L, Fang T, Zhang T, Jiang J, Li H. Niu X, et al. Front Oncol. 2020 Dec 17;10:590352. doi: 10.3389/fonc.2020.590352. eCollection 2020. Front Oncol. 2020. PMID: 33392085 Free PMC article. - Development of a 3 RNA Binding Protein Signature for Predicting Prognosis and Treatment Response for Glioblastoma Multiforme.
Sun R, Pan Y, Mu L, Ma Y, Shen H, Long Y. Sun R, et al. Front Genet. 2021 Oct 18;12:768930. doi: 10.3389/fgene.2021.768930. eCollection 2021. Front Genet. 2021. PMID: 34733320 Free PMC article. - Identification of a multidimensional transcriptome signature for survival prediction of postoperative glioblastoma multiforme patients.
Gao WZ, Guo LM, Xu TQ, Yin YH, Jia F. Gao WZ, et al. J Transl Med. 2018 Dec 20;16(1):368. doi: 10.1186/s12967-018-1744-8. J Transl Med. 2018. PMID: 30572911 Free PMC article.
Cited by
- Angiogenesis-Related Gene Signature-Derived Risk Score for Glioblastoma: Prospects for Predicting Prognosis and Immune Heterogeneity in Glioblastoma.
Wang G, Hu JQ, Liu JY, Zhang XM. Wang G, et al. Front Cell Dev Biol. 2022 Mar 18;10:778286. doi: 10.3389/fcell.2022.778286. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35372355 Free PMC article. - Large-Scale Analysis Reveals Gene Signature for Survival Prediction in Primary Glioblastoma.
Prasad B, Tian Y, Li X. Prasad B, et al. Mol Neurobiol. 2020 Dec;57(12):5235-5246. doi: 10.1007/s12035-020-02088-w. Epub 2020 Sep 1. Mol Neurobiol. 2020. PMID: 32875483 Free PMC article. - Analysis of the prognostic significance of solute carrier (SLC) family 39 genes in breast cancer.
Liu L, Yang J, Wang C. Liu L, et al. Biosci Rep. 2020 Aug 28;40(8):BSR20200764. doi: 10.1042/BSR20200764. Biosci Rep. 2020. PMID: 32744318 Free PMC article. - A comprehensive prognostic signature for glioblastoma patients based on transcriptomics and single cell sequencing.
Fan F, Zhang H, Dai Z, Zhang Y, Xia Z, Cao H, Yang K, Hu S, Guo Y, Ding F, Cheng Q, Zhang N. Fan F, et al. Cell Oncol (Dordr). 2021 Aug;44(4):917-935. doi: 10.1007/s13402-021-00612-1. Epub 2021 Jun 17. Cell Oncol (Dordr). 2021. PMID: 34142341 - HOXC10 upregulation confers resistance to chemoradiotherapy in ESCC tumor cells and predicts poor prognosis.
Suo D, Wang Z, Li L, Chen Q, Zeng T, Liu R, Yun J, Guan XY, Li Y. Suo D, et al. Oncogene. 2020 Aug;39(32):5441-5454. doi: 10.1038/s41388-020-1375-4. Epub 2020 Jun 25. Oncogene. 2020. PMID: 32587398
References
- Wen PY, Reardon DA Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12:69–70. - PubMed
LinkOut - more resources
Full Text Sources