Rhopalocnemis phalloides has one of the most reduced and mutated plastid genomes known - PubMed (original) (raw)
Rhopalocnemis phalloides has one of the most reduced and mutated plastid genomes known
Mikhail I Schelkunov et al. PeerJ. 2019.
Abstract
Although most plant species are photosynthetic, several hundred species have lost the ability to photosynthesize and instead obtain nutrients via various types of heterotrophic feeding. Their plastid genomes markedly differ from the plastid genomes of photosynthetic plants. In this work, we describe the sequenced plastid genome of the heterotrophic plant Rhopalocnemis phalloides, which belongs to the family Balanophoraceae and feeds by parasitizing other plants. The genome is highly reduced (18,622 base pairs vs. approximately 150 kbp in autotrophic plants) and possesses an extraordinarily high AT content, 86.8%, which is inferior only to AT contents of plastid genomes of Balanophora, a genus from the same family. The gene content of this genome is quite typical of heterotrophic plants, with all of the genes related to photosynthesis having been lost. The remaining genes are notably distorted by a high mutation rate and the aforementioned AT content. The high AT content has led to sequence convergence between some of the remaining genes and their homologs from AT-rich plastid genomes of protists. Overall, the plastid genome of R. phalloides is one of the most unusual plastid genomes known.
Keywords: AT content; Chloroplast genomes; GC content; Heterotrophic plants; Parasitic plants; Plastid genomes.
© 2019 Schelkunov et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
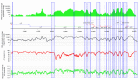
Figure 1. Map of the Rhopalocnemis phalloides plastid genome showing various features.
The circular-mapping plastid genome is represented linearly for convenience. Green arrows are rRNA-coding genes, red arrows are ribosomal protein-coding genes and blue arrows are genes coding proteins with other functions. Gray arcs represent splicing. Blue columns show non-coding regions.
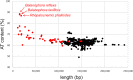
Figure 2. AT content and lengths of the plastid genomes of Embryophyta.
Red dots denote completely heterotrophic plants and black dots mixotrophic and completely autotrophic.
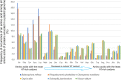
Figure 3. Amino acid frequencies in the plastid proteins of Rhopalocnemis phalloides and Balanophora reflexa are affected by the high AT content.
Figure 4. The radial cladogram of rrn16 from Embryophyta and SAR.
Branches with bootstrap support below 70 are collapsed. The group of SAR with Balanophoraceae is colored orange, while Embryophyta without Balanophoraceae are colored green. The placement of Balanophoraceae within SAR is indicated by pink.
Figure 5. Evolutionary parameters of the phylogeny of Santalales.
Arabidopsis thaliana, used as the outgroup, is not shown. The total length of the alignment, used for the analysis, was 3,363 bp after removal of poorly aligned regions by Gblocks. * dN/dS on this branch cannot be calculated owing to a very small dS value.
Similar articles
- Invited Review Beyond parasitic convergence: unravelling the evolution of the organellar genomes in holoparasites.
Sanchez-Puerta MV, Ceriotti LF, Gatica-Soria LM, Roulet ME, Garcia LE, Sato HA. Sanchez-Puerta MV, et al. Ann Bot. 2023 Nov 30;132(5):909-928. doi: 10.1093/aob/mcad108. Ann Bot. 2023. PMID: 37503831 Free PMC article. Review. - Genomic comparison of non-photosynthetic plants from the family Balanophoraceae with their photosynthetic relatives.
Schelkunov MI, Nuraliev MS, Logacheva MD. Schelkunov MI, et al. PeerJ. 2021 Aug 31;9:e12106. doi: 10.7717/peerj.12106. eCollection 2021. PeerJ. 2021. PMID: 34540375 Free PMC article. - Mycoheterotrophic Epirixanthes (Polygalaceae) has a typical angiosperm mitogenome but unorthodox plastid genomes.
Petersen G, Darby H, Lam VKY, Pedersen HÆ, Merckx VSFT, Zervas A, Seberg O, Graham SW. Petersen G, et al. Ann Bot. 2019 Nov 15;124(5):791-807. doi: 10.1093/aob/mcz114. Ann Bot. 2019. PMID: 31346602 Free PMC article. - The loss of photosynthetic pathways in the plastid and nuclear genomes of the non-photosynthetic mycoheterotrophic eudicot Monotropa hypopitys.
Ravin NV, Gruzdev EV, Beletsky AV, Mazur AM, Prokhortchouk EB, Filyushin MA, Kochieva EZ, Kadnikov VV, Mardanov AV, Skryabin KG. Ravin NV, et al. BMC Plant Biol. 2016 Nov 16;16(Suppl 3):238. doi: 10.1186/s12870-016-0929-7. BMC Plant Biol. 2016. PMID: 28105941 Free PMC article. - Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists.
Hadariová L, Vesteg M, Hampl V, Krajčovič J. Hadariová L, et al. Curr Genet. 2018 Apr;64(2):365-387. doi: 10.1007/s00294-017-0761-0. Epub 2017 Oct 12. Curr Genet. 2018. PMID: 29026976 Review.
Cited by
- Phylogenetic position and plastid genome structure of Vietorchis, a mycoheterotrophic genus of Orchidaceae (subtribe Orchidinae) endemic to Vietnam.
Samigullin TH, Logacheva MD, Averyanov LV, Zeng SJ, Fu LF, Nuraliev MS. Samigullin TH, et al. Front Plant Sci. 2024 May 24;15:1393225. doi: 10.3389/fpls.2024.1393225. eCollection 2024. Front Plant Sci. 2024. PMID: 38855461 Free PMC article. - Invited Review Beyond parasitic convergence: unravelling the evolution of the organellar genomes in holoparasites.
Sanchez-Puerta MV, Ceriotti LF, Gatica-Soria LM, Roulet ME, Garcia LE, Sato HA. Sanchez-Puerta MV, et al. Ann Bot. 2023 Nov 30;132(5):909-928. doi: 10.1093/aob/mcad108. Ann Bot. 2023. PMID: 37503831 Free PMC article. Review. - Extreme plastomes in holoparasitic Balanophoraceae are not the norm.
Kim W, Lautenschläger T, Bolin JF, Rees M, Nzuzi A, Zhou R, Wanke S, Jost M. Kim W, et al. BMC Genomics. 2023 Jun 15;24(1):330. doi: 10.1186/s12864-023-09422-1. BMC Genomics. 2023. PMID: 37322447 Free PMC article. - Cytonuclear coevolution in a holoparasitic plant with highly disparate organellar genomes.
Ceriotti LF, Gatica-Soria L, Sanchez-Puerta MV. Ceriotti LF, et al. Plant Mol Biol. 2022 Aug;109(6):673-688. doi: 10.1007/s11103-022-01266-9. Epub 2022 Mar 31. Plant Mol Biol. 2022. PMID: 35359176
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
- Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution. 2014;31(12):3095–3112. doi: 10.1093/molbev/msu252. - DOI - PubMed
Grants and funding
The work was funded by a Russian Foundation for Basic Research grant (No. 16-34-01003) and a budgetary subsidy to the Institute for Information Transmission Problems (No. 0053-2019-0005). The work of Maxim Segreevich Nuraliev was carried out in accordance with a Government order for the Lomonosov Moscow State University (project No. AAAA-A16-116021660105-3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Miscellaneous