The human body at cellular resolution: the NIH Human Biomolecular Atlas Program - PubMed (original) (raw)
The human body at cellular resolution: the NIH Human Biomolecular Atlas Program
HuBMAP Consortium. Nature. 2019 Oct.
Abstract
Transformative technologies are enabling the construction of three-dimensional maps of tissues with unprecedented spatial and molecular resolution. Over the next seven years, the NIH Common Fund Human Biomolecular Atlas Program (HuBMAP) intends to develop a widely accessible framework for comprehensively mapping the human body at single-cell resolution by supporting technology development, data acquisition, and detailed spatial mapping. HuBMAP will integrate its efforts with other funding agencies, programs, consortia, and the biomedical research community at large towards the shared vision of a comprehensive, accessible three-dimensional molecular and cellular atlas of the human body, in health and under various disease conditions.
Conflict of interest statement
M.P.S. is a cofounder and on the scientific advisory board of Personalis, Filtircine, SensOmics, Qbio, January, Mirvie, Oralome and Proteus. He is also on the scientific advisory board (SAB) of Genapsys and Jupiter and on the advisory board of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). P.V.K. serves on the SAB to Celsius Therapeutics. A.R. is a member of the SAB of ThermoFisher Scientific and Syros Pharmaceuticals and a founder and an equity holder of Celsius Therapeutics. A.R. holds various patents and has patent filings in the areas of single-cell and spatial genomic technologies, and is a member of the advisory council of the National Human Genome Research Institute (NHGRI). C.K. is a co-founder of Ocean Genomics. P.Y. is a co-founder, paid consultant, director, and equity holder of Ultivue and NuProbe Global and holds several patent filings in the areas of single-cell and spatial genomic technologies. N.G. is a co-founder and equity owner of Datavisyn. R.F.M. is a cofounder and board member of Quantitative Medicine and is on the Advisory Board of Predictive Oncology and the Scientific Advisory Board of the Morgridge Institute for Research. The other authors declare no competing interests.
Figures
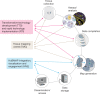
Fig. 1. The HubMAP consortium.
The TMCs will collect tissue samples and generate spatially resolved, single-cell data. Groups involved in TTD and RTI initiatives will develop emerging and more developed technologies, respectively; in later years, these will be implemented at scale. Data from all groups will be rendered useable for the biomedical community by the HuBMAP integration, visualization and engagement (HIVE) teams. The groups will collaborate closely to iteratively refine the atlas as it is gradually realized.
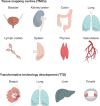
Fig. 2. Key tissues and organs initially analysed by the consortium.
Using innovative, production-grade (‘shovel ready’) technologies, HuBMAP TMCs will generate data for single-cell, three-dimensional maps of various human tissues. In parallel, TTD projects (and later RTI projects) will refine assays and analysis tools on a largely distinct set of human tissues. Samples from individuals of both sexes and different ages will be studied. The range of tissues will be expanded throughout the program.
Fig. 3. Map generation and assembly across cellular and spatial scales.
HuBMAP aims to produce an atlas in which users can refer to a histological slide from a specific part of an organ and, in any given cell, understand its contents on multiple ’omic levels—genomic, epigenomic, transcriptomic, proteomic, and/or metabolomic. To achieve these ends, centres will apply a combination of imaging, ’omics and mass spectrometry techniques to specimens collected in a reproducible manner from specific sites in the body. These data will be then be integrated to arrive at a high-resolution, high-content three-dimensional map for any given tissue. To ensure inter-individual differences will not be confounded with collection heterogeneity, a robust CCF will be developed.
Similar articles
- COVID, racism, China: three tests for the next NIH leader.
[No authors listed] [No authors listed] Nature. 2021 Oct;598(7881):385-386. doi: 10.1038/d41586-021-02842-7. Nature. 2021. PMID: 34667292 No abstract available. - China. After long march, scientists create 'Chinese NIH'.
Jiao L. Jiao L. Science. 2010 Jan 8;327(5962):132. doi: 10.1126/science.327.5962.132-a. Science. 2010. PMID: 20056862 No abstract available. - Human BioMolecular Atlas Program (HuBMAP): 3D Human Reference Atlas construction and usage.
Börner K, Blood PD, Silverstein JC, Ruffalo M, Satija R, Teichmann SA, Pryhuber GJ, Misra RS, Purkerson JM, Fan J, Hickey JW, Molla G, Xu C, Zhang Y, Weber GM, Jain Y, Qaurooni D, Kong Y; HRA Team; Bueckle A, Herr BW 2nd. Börner K, et al. Nat Methods. 2025 Apr;22(4):845-860. doi: 10.1038/s41592-024-02563-5. Epub 2025 Mar 13. Nat Methods. 2025. PMID: 40082611 Free PMC article. - Advances and prospects for the Human BioMolecular Atlas Program (HuBMAP).
Jain S, Pei L, Spraggins JM, Angelo M, Carson JP, Gehlenborg N, Ginty F, Gonçalves JP, Hagood JS, Hickey JW, Kelleher NL, Laurent LC, Lin S, Lin Y, Liu H, Naba A, Nakayasu ES, Qian WJ, Radtke A, Robson P, Stockwell BR, Van de Plas R, Vlachos IS, Zhou M; HuBMAP Consortium; Börner K, Snyder MP. Jain S, et al. Nat Cell Biol. 2023 Aug;25(8):1089-1100. doi: 10.1038/s41556-023-01194-w. Epub 2023 Jul 19. Nat Cell Biol. 2023. PMID: 37468756 Free PMC article. Review. - Disproportionally low funding for trauma research by the National Institutes of Health: A call for a National Institute of Trauma.
Glass NE, Riccardi J, Farber NI, Bonne SL, Livingston DH. Glass NE, et al. J Trauma Acute Care Surg. 2020 Jan;88(1):25-32. doi: 10.1097/TA.0000000000002461. J Trauma Acute Care Surg. 2020. PMID: 31389923 Review.
Cited by
- Giotto: a toolbox for integrative analysis and visualization of spatial expression data.
Dries R, Zhu Q, Dong R, Eng CL, Li H, Liu K, Fu Y, Zhao T, Sarkar A, Bao F, George RE, Pierson N, Cai L, Yuan GC. Dries R, et al. Genome Biol. 2021 Mar 8;22(1):78. doi: 10.1186/s13059-021-02286-2. Genome Biol. 2021. PMID: 33685491 Free PMC article. - PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements.
Seo J, Sim Y, Kim J, Kim H, Cho I, Nam H, Yoon YG, Chang JB. Seo J, et al. Nat Commun. 2022 May 5;13(1):2475. doi: 10.1038/s41467-022-30168-z. Nat Commun. 2022. PMID: 35513404 Free PMC article. - 3D virtual reality vs. 2D desktop registration user interface comparison.
Bueckle A, Buehling K, Shih PC, Börner K. Bueckle A, et al. PLoS One. 2021 Oct 27;16(10):e0258103. doi: 10.1371/journal.pone.0258103. eCollection 2021. PLoS One. 2021. PMID: 34705835 Free PMC article. - Childhood rare lung disease in the 21st century: "-omics" technology advances accelerating discovery.
Vece TJ, Wambach JA, Hagood JS. Vece TJ, et al. Pediatr Pulmonol. 2020 Jul;55(7):1828-1837. doi: 10.1002/ppul.24809. Pediatr Pulmonol. 2020. PMID: 32533908 Free PMC article. Review. - Transcriptomic data meta-analysis reveals common and injury model specific gene expression changes in the regenerating zebrafish heart.
Botos MA, Arora P, Chouvardas P, Mercader N. Botos MA, et al. Sci Rep. 2023 Apr 3;13(1):5418. doi: 10.1038/s41598-023-32272-6. Sci Rep. 2023. PMID: 37012284 Free PMC article.
References
- Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat. Rev. Genet. 2015;16:716–726. - PubMed
Publication types
MeSH terms
Grants and funding
- OT2 OD026675/OD/NIH HHS/United States
- 22231/CRUK_/Cancer Research UK/United Kingdom
- OT2 OD026663/OD/NIH HHS/United States
- U54 DK120058/DK/NIDDK NIH HHS/United States
- UG3 HL145600/HL/NHLBI NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- OT2 OD026671/OD/NIH HHS/United States
- OT2 OD026682/OD/NIH HHS/United States
- U54 HL145608/HL/NHLBI NIH HHS/United States
- UG3 HL145593/HL/NHLBI NIH HHS/United States
- OT2 OD026673/OD/NIH HHS/United States
- OT2 OD026677/OD/NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- UG3 HL145623/HL/NHLBI NIH HHS/United States
- UG3 HL145609/HL/NHLBI NIH HHS/United States
- U54 HL145611/HL/NHLBI NIH HHS/United States
- U54 HG010426/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources