Unique structural features in an Nramp metal transporter impart substrate-specific proton cotransport and a kinetic bias to favor import - PubMed (original) (raw)
Unique structural features in an Nramp metal transporter impart substrate-specific proton cotransport and a kinetic bias to favor import
Aaron T Bozzi et al. J Gen Physiol. 2019.
Abstract
Natural resistance-associated macrophage protein (Nramp) transporters enable uptake of essential transition metal micronutrients in numerous biological contexts. These proteins are believed to function as secondary transporters that harness the electrochemical energy of proton gradients by "coupling" proton and metal transport. Here we use the Deinococcus radiodurans (Dra) Nramp homologue, for which we have determined crystal structures in multiple conformations, to investigate mechanistic details of metal and proton transport. We untangle the proton-metal coupling behavior of DraNramp into two distinct phenomena: ΔpH stimulation of metal transport rates and metal stimulation of proton transport. Surprisingly, metal type influences substrate stoichiometry, leading to manganese-proton cotransport but cadmium uniport, while proton uniport also occurs. Additionally, a physiological negative membrane potential is required for high-affinity metal uptake. To begin to understand how Nramp's structure imparts these properties, we target a conserved salt-bridge network that forms a proton-transport pathway from the metal-binding site to the cytosol. Mutations to this network diminish voltage and ΔpH dependence of metal transport rates, alter substrate selectivity, perturb or eliminate metal-stimulated proton transport, and erode the directional bias favoring outward-to-inward metal transport under physiological-like conditions. Thus, this unique salt-bridge network may help Nramp-family transporters maximize metal uptake and reduce deleterious back-transport of acquired metals. We provide a new mechanistic model for Nramp proton-metal cotransport and propose that functional advantages may arise from deviations from the traditional model of symport.
© 2019 Bozzi et al.
Figures
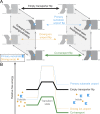
Figure 1.
Kinetic model of canonical symport. (A) Transport cycle diagram illustrating all possible binding/unbinding/transport events for a canonical symporter. (B) Free energy diagrams for transport events. For a tightly coupled (canonical) symporter, essentially insurmountable kinetic barriers in the protein’s free energy landscape prevent uniport events (dashed lines). Thus, only the empty or fully loaded transporter (solid lines) can efficiently convert from outward-open to inward-open (or vice versa). The combined electrochemical gradients of the primary substrate and the driving ions determine the net direction of cotransport, with a typical physiological situation for symport displayed here (higher concentration of primary substrate inside and higher concentration of driving ion outside).
Figure 2.
Metal transport shows strong voltage dependence. (A) Schematic of proteoliposome assay for metal transport at variable membrane potentials (ΔΨ) established by using K+ gradients and valinomycin. The metal-sensitive dye Fura-2 was used to detect metal uptake. No pH gradient was used in these experiments. (B) Representative time traces (n = 8) of Cd2+, Mn2+, and Co2+ transport at different ΔΨs; only Cd2+ is transported at ΔΨ = 0 mV. After recording a baseline, the indicated external metal concentration (750 µM for these and all subsequent traces unless otherwise indicated) and valinomycin were added, and transport was monitored for 5–10 min. (C) Average initial transport rate ± SEM (n = 4) versus membrane potential. Each metal except Cd2+ has a characteristic threshold negative voltage for transport to occur: −10 mV for Mn2+, −50 mV for Fe2+ and Zn2+, and −80 mV for Co2+. (D) Average initial transport rate ± SEM (n = 2–3) versus metal concentration at ΔΨ = −150 mV. Comparison of C and D (table inset) shows that higher-magnitude threshold voltages correlate with _K_m, indicating that voltage dependence of transport rate may be enforced through a metal-binding step. Errors in _K_m and _V_max encompass the uncertainty of fit (shown as curved lines) to data. (E) Time course of Co2+ uptake at pH 7.0 and variable [KCl] shows that high extracellular [KCl] reduces Co2+ uptake in E. coli. (F) Complementary single-cysteine reporters to assess conformational cycling of DraNramp. (G) Fraction of modified cysteine versus NEM concentration at pH 7. Inward (A53C) and outward (A61C) reporters were both labeled, even at high [KCl]out that depresses Co2+ uptake. Conformational locking thus cannot explain reduced Co2+ transport by DraNramp under these conditions. Data in E and G are averages ± SEM (n = 4). See also Fig. S1.
Figure 3.
Acidic external pH and favorable outside-to-inside pH gradients stimulate metal transport. (A) Schematic for assessing effect of varying external pH on metal transport in proteoliposomes with internal pH 7. K+ gradients and valinomycin were used to set ΔΨ = 0 or −60 mV. A strong external buffer was added to adjust the pH to the indicated value when metal and valinomycin were added after the baseline read. (B and C) Representative time traces (n ≥ 4) of 750 µM Cd2+ (B) or Mn2+ (C) uptake into proteoliposomes. Acidic-outside ΔpH accelerated Cd2+ and Mn2+ transport (at ΔΨ = −60 mV) and enabled Mn2+ uptake when ΔΨ = 0. (D) Schematic for discriminating between effect of low pH and ΔpH on metal transport rate by varying internal and external pH. (E and F) Initial metal uptake rates show that Cd2+ uptake (E) was accelerated by a favorable ΔpH, while Mn2+ uptake (F) was faster in the presence of lower external pH and further accelerated by a favorable ΔpH. Data are averages ± SEM (n ≥ 3). See also Fig. S2.
Figure 4.
Voltage and pH gradients govern metal transport kinetics. (A and B) Dose–response curves for Cd2+ (A) and Mn2+ (B) transport at different external pH and ΔΨ values in proteoliposomes with internal pH = 7. Data are averages ± SEM (n = 3). Errors in _K_m and _V_max reported in the inset table encompass the uncertainty of the fit (shown as curved lines) to the data.
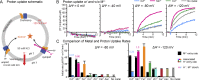
Figure 5.
Proton cotransport is metal specific. (A) Schematic for detecting H+ influx into proteoliposomes at various ΔΨ values, pH 7.0 on both sides, in the absence or presence of 750 µM metal substrate. The setup was identical to that used in Fig. 2 (A and B), except the pH-sensitive dye BCECF replaced the metal-sensitive dye Fura-2. (B) Representative time traces (n = 8) of H+ uptake into proteoliposomes. Negative ΔΨ drove DraNramp H+ import. Mn2+ and Co2+ stimulated H+ entry, while Cd2+ did not and instead reduced H+ influx at ΔΨ = −120 mV. (C) Initial metal (black) and H+ (color) uptake rates show that Mn2+, Fe2+, and Co2+ transport stimulated H+ influx above its basal no-metal rate, while Cd2+ and Zn2+ transport did not. Stoichiometry ratios (numbers above bars) were calculated for Cd2+ and Mn2+ and were ∼1:1 for Mn2+:H+ transport in the presence of metal. Data are averages ± SEM (n ≥ 4). See also Fig. S3.
Figure 6.
A network of highly conserved charged and protonatable residues adjoins the metal-binding site. (A) Crystal structure of DraNramp in an outward-open conformation bound to Mn2+ (magenta sphere; PDB accession no. 6BU5; Bozzi et al., 2019). TMs 1, 5, 6, and 10 are gold; TMs 2, 7, and 11 are gray; and TMs 3, 4, 8, and 9 are blue. (B) D56, N59, M230, and the A53 carbonyl coordinate Mn2+ in the outward-open state, along with two waters (not depicted). (C) View from the plane of the membrane of a network consisting of E134, H232, D131, R353, R352, and E124 that extends ∼20 Å from D56 to the cytosol. TMs 8 and 4–5, in front of TMs 3 and 9, respectively, were removed for clarity. (D) Sequence logos from an alignment of 6,878 Nramp sequences. All 10 mutated residues (numbers above) are highly conserved. Canonical helix-breaking motifs at the metal-binding site are DPGN in TM1 and MPH in TM6. TM6’s H237 is a glycine in many fungal homologues. The second TM9 arginine (R353) varies in location (*) due to an extra helical turn in many homologues; this insertion contains a third arginine in some homologues. See also Fig. S4.
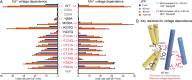
Figure 7.
Mutations to the conserved salt-bridge network perturb voltage dependence of metal transport rate. (A) Average initial metal uptake rates ± SEM (n ≥ 4) for DraNramp mutants at different ΔΨ (0 to −120 mV, colored bars). [M2+] was 750 µM, and the pH was 7 on both sides of the membrane. The fold increase in transport rate is indicated (cyan, 0 to −120 mV for Cd2+; pink, −40 to −120 mV for Mn2+). Mutations to E124, D131, E134, R352, and R353 (red) reduced the ΔΨ dependence of Cd2+ and Mn2+ transport rates across the range of tested voltages, such that they exceeded WT at low-magnitude ΔΨ but lagged WT at physiological large-magnitude ΔΨ. Mutations to N59, M230, H232, and H237 (black) retained WT-like ΔΨ dependence, and D56 mutants (gray) eliminated all metal transport. (B) Network schematic illustrates clustering of residues most important for ΔΨ dependence (red). See also Fig. S5 for representative time traces of these results and Fig. S6 for dependence of in vivo Co2+ uptake rates on external [K+].
Figure 8.
Mutations to conserved network reduce or eliminate ΔpH stimulation of Mn2+ transport and perturb or eliminate H+ cotransport. (A) Average initial Mn2+ uptake rates ± SEM (n ≥ 4) in proteoliposomes at various external pHs, internal pH = 7, and ΔΨ = −60 mV, as in Fig. 3 (A–C). [Mn2+] was 750 µM. (B) Schematic shows proximity to binding site of residues where mutations fully eliminate the ΔpH stimulation effect (red), while more distant mutations within the salt-bridge network still decrease the ΔpH effect (orange). See also Fig. S7 for representative time traces of these results. (C) Average initial metal-stimulated H+ transport rates ± SEM (n ≥ 4) at ΔΨ = −120 mV with pH 7.0 on both sides of the membrane, in the absence or presence of 750 µM M2+ as in Fig. 5. (D) Schematic shows that protonatable residues in immediate vicinity of D56 are essential for metal-stimulated H+ transport, while the entire network affects proton-metal cotransport behavior. Interestingly, D131 and E134 mutants retain some ΔpH stimulation while eliminating metal-stimulated H+ cotransport, while M230A shows the opposite pattern. Mutations to the salt-bridge network residues furthest from D56 increase rate of H+ transport without metal but have variable effects on metal stimulation. See also Fig. S8 for representative time traces of these results and Fig. S9.
Figure 9.
Perturbations to the conserved salt-bridge network alter metal selectivity and increase _K_m for Mn2+. (A) Average initial Ca2+ transport rates ± SEM (n ≥ 4) at −120 mV with pH 7.0 on both sides of the membrane and 750 µM Ca2+. (B) Schematic of mutation locations. M230A, or R353A, 15 Å away; both increased Ca2+ transport. D56 and N59 mutations eliminated Ca2+ uptake, and other mutants were similar to WT. (C and D) Dose–response curves of Cd2+ (C) or Mn2+ (D) transport at −150 mV with pH 7.0 on both sides of the membrane for a subset of mutants that reduce or eliminate ΔΨ dependence, ΔpH stimulation, or metal-dependent proton transport. Data are averages ± SEM (n ≥ 2). The resulting transport kinetic values (middle) show significant overlap for Cd2+ transport but wider separation for the physiological substrate Mn2+. M230A is the only mutation that impaired Cd2+ transport more severely than Mn2+ transport. Errors in _K_m and _V_max encompass the uncertainty of the fit to the data. See also Fig. S10 for representative time traces of the data in A.
Figure 10.
Mutations to the salt-bridge network enable deleterious Mn2+ back transport. (A) Schematic for isolating metal-transport activity from the ∼50% inside-out transporters (Bozzi et al., 2019). Only outside-out transporters are incapacitated by A61C modification with MTSET. (B) Average initial transport rates ± SEM (n = 4) of MTSET-treated or untreated proteoliposomes at ΔΨ = −50 mV (left) or +50 mV (right; note that the axis scales are different). pH = 7 on both sides of the membrane, and [M2+] = 1.5 mM. MTSET essentially eliminated WT-like’s Mn2+ transport. In contrast, D131N, E134A, and R352A retained significant Mn2+ transport, likely reflecting increased activity from inside-out transporters. In addition, MTSET treatment eliminated more Mn2+ transport than Cd2+ transport, which did not require H+ cotransport (Fig. 5). (C) A61C liposome assay mimics cellular context for DraNramp back transport. Metal influx into MTSET-treated proteoliposomes occurs down a concentration gradient but against a ΔΨ > 0, just as metal efflux would in vivo. See also Fig. S11 for representative time traces of these results.
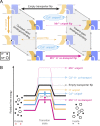
Figure 11.
Kinetic model of DraNramp activity. (A) Transport cycle diagram including all possible binding/unbinding events as well as all observed (solid lines) and hypothetical (dashed lines) transport events for DraNramp. While metal transport always requires bulk conformational change, proton uniport occurs through the outward-open state (Bozzi et al., 2019). Cd2+ uniport (horizontal cycles) could occur in the presence or absence of a bound proton. (B) Simplified, hypothetical free energy diagrams for DraNramp transport events in a typical physiological context of higher [H+] outside, higher [M2+] inside, and a moderate negative-inside ΔΨ. Proton cotransport may significantly reduce the barrier to Mn2+ transport, while Cd2+ uniport may instead have the lowest barrier under physiological conditions. Voltage affects both the magnitude of the energetic barriers for metal transport as well as the relative ΔG for transport, while ΔpH affects the energetic barrier and relative ΔG depending on the extent of thermodynamic coupling between metal and proton transport. The much faster rates seen for metal forward-transport over back-transport imply asymmetric kinetic barriers (and thus likely additional stable intermediate states), which are not shown in this model.
Figure 12.
Proposed DraNramp proton-metal cotransport mechanism. (A) Proposed mechanism of proton-metal cotransport. Left: D56 protonation optimizes the metal-binding site via hydrogen bonding of D56 to the N59 carbonyl oxygen, providing a better metal-binding ligand than the amide nitrogen (as shown in B). Center: Metal substrate binds, displacing the D56 proton, which passes to D131, with H232 and E134 stabilizing a high-energy transition state and triggering bulk conformational change. Right: The proton exits to the cytosol via the TM3/TM9 salt-bridge network, while the metal is released into the cytosolic vestibule. (B) Detailed view of how protonation of D56 could alter the metal-binding site. (C and D) Cd2+ uniport perhaps occurs due to a monodentate interaction with D56, enabling proton retention. Bidentate binding of D56 by Mn2+ requires deprotonation, passing the proton to D131. See also Table S1.
Comment in
- Unconventional transport of metal ions and protons by Nramps.
Rudnick G. Rudnick G. J Gen Physiol. 2019 Dec 2;151(12):1339-1342. doi: 10.1085/jgp.201912464. Epub 2019 Nov 5. J Gen Physiol. 2019. PMID: 31690583 Free PMC article.
Similar articles
- Transmembrane helix 6b links proton and metal release pathways and drives conformational change in an Nramp-family transition metal transporter.
Bozzi AT, McCabe AL, Barnett BC, Gaudet R. Bozzi AT, et al. J Biol Chem. 2020 Jan 31;295(5):1212-1224. doi: 10.1074/jbc.RA119.011336. Epub 2019 Dec 27. J Biol Chem. 2020. PMID: 31882536 Free PMC article. - Molecular Mechanism of Nramp-Family Transition Metal Transport.
Bozzi AT, Gaudet R. Bozzi AT, et al. J Mol Biol. 2021 Aug 6;433(16):166991. doi: 10.1016/j.jmb.2021.166991. Epub 2021 Apr 16. J Mol Biol. 2021. PMID: 33865868 Free PMC article. Review. - Structures in multiple conformations reveal distinct transition metal and proton pathways in an Nramp transporter.
Bozzi AT, Zimanyi CM, Nicoludis JM, Lee BK, Zhang CH, Gaudet R. Bozzi AT, et al. Elife. 2019 Feb 4;8:e41124. doi: 10.7554/eLife.41124. Elife. 2019. PMID: 30714568 Free PMC article. - Crystal Structure and Conformational Change Mechanism of a Bacterial Nramp-Family Divalent Metal Transporter.
Bozzi AT, Bane LB, Weihofen WA, Singharoy A, Guillen ER, Ploegh HL, Schulten K, Gaudet R. Bozzi AT, et al. Structure. 2016 Dec 6;24(12):2102-2114. doi: 10.1016/j.str.2016.09.017. Epub 2016 Nov 10. Structure. 2016. PMID: 27839948 Free PMC article. - Recent progress in structure-function analyses of Nramp proton-dependent metal-ion transporters.
Courville P, Chaloupka R, Cellier MF. Courville P, et al. Biochem Cell Biol. 2006 Dec;84(6):960-78. doi: 10.1139/o06-193. Biochem Cell Biol. 2006. PMID: 17215883 Review.
Cited by
- Ion and lipid orchestration of secondary active transport.
Drew D, Boudker O. Drew D, et al. Nature. 2024 Feb;626(8001):963-974. doi: 10.1038/s41586-024-07062-3. Epub 2024 Feb 28. Nature. 2024. PMID: 38418916 Review. - Iron transport pathways in the human malaria parasite Plasmodium falciparum revealed by RNA-sequencing.
Wunderlich J, Kotov V, Votborg-Novél L, Ntalla C, Geffken M, Peine S, Portugal S, Strauss J. Wunderlich J, et al. Front Cell Infect Microbiol. 2024 Nov 7;14:1480076. doi: 10.3389/fcimb.2024.1480076. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39575308 Free PMC article. - Nramp: Deprive and conquer?
Cellier MFM. Cellier MFM. Front Cell Dev Biol. 2022 Oct 13;10:988866. doi: 10.3389/fcell.2022.988866. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313567 Free PMC article. - Transmembrane helix 6b links proton and metal release pathways and drives conformational change in an Nramp-family transition metal transporter.
Bozzi AT, McCabe AL, Barnett BC, Gaudet R. Bozzi AT, et al. J Biol Chem. 2020 Jan 31;295(5):1212-1224. doi: 10.1074/jbc.RA119.011336. Epub 2019 Dec 27. J Biol Chem. 2020. PMID: 31882536 Free PMC article. - Molecular Mechanism of Nramp-Family Transition Metal Transport.
Bozzi AT, Gaudet R. Bozzi AT, et al. J Mol Biol. 2021 Aug 6;433(16):166991. doi: 10.1016/j.jmb.2021.166991. Epub 2021 Apr 16. J Mol Biol. 2021. PMID: 33865868 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources