MGnify: the microbiome analysis resource in 2020 - PubMed (original) (raw)
. 2020 Jan 8;48(D1):D570-D578.
doi: 10.1093/nar/gkz1035.
Alexandre Almeida 1 2, Martin Beracochea 1, Miguel Boland 1, Josephine Burgin 1, Guy Cochrane 1, Michael R Crusoe 3, Varsha Kale 1, Simon C Potter 1, Lorna J Richardson 1, Ekaterina Sakharova 1, Maxim Scheremetjew 1, Anton Korobeynikov 4, Alex Shlemov 4, Olga Kunyavskaya 4, Alla Lapidus 4, Robert D Finn 1
Affiliations
- PMID: 31696235
- PMCID: PMC7145632
- DOI: 10.1093/nar/gkz1035
MGnify: the microbiome analysis resource in 2020
Alex L Mitchell et al. Nucleic Acids Res. 2020.
Abstract
MGnify (http://www.ebi.ac.uk/metagenomics) provides a free to use platform for the assembly, analysis and archiving of microbiome data derived from sequencing microbial populations that are present in particular environments. Over the past 2 years, MGnify (formerly EBI Metagenomics) has more than doubled the number of publicly available analysed datasets held within the resource. Recently, an updated approach to data analysis has been unveiled (version 5.0), replacing the previous single pipeline with multiple analysis pipelines that are tailored according to the input data, and that are formally described using the Common Workflow Language, enabling greater provenance, reusability, and reproducibility. MGnify's new analysis pipelines offer additional approaches for taxonomic assertions based on ribosomal internal transcribed spacer regions (ITS1/2) and expanded protein functional annotations. Biochemical pathways and systems predictions have also been added for assembled contigs. MGnify's growing focus on the assembly of metagenomic data has also seen the number of datasets it has assembled and analysed increase six-fold. The non-redundant protein database constructed from the proteins encoded by these assemblies now exceeds 1 billion sequences. Meanwhile, a newly developed contig viewer provides fine-grained visualisation of the assembled contigs and their enriched annotations.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
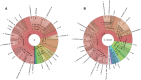
Figure 1.
Krona plots showing ITS-based taxonomic analysis results for sequencing run ERR2237853 from agricultural soil. Panel A shows the analysis results obtained using the UNITE database and B shows those produced with ITSoneDB. These plots are integrated into the taxonomy tab within the analysis results section,
https://www.ebi.ac.uk/metagenomics/analyses/MGYA00383253#taxonomic
.
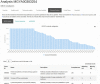
Figure 2.
Screenshot showing the percentage completion for a series of KEGG pathways based on the presence of KEGG orthologues,
https://www.ebi.ac.uk/metagenomics/analyses/MGYA00383254#path-systems
.
Figure 3.
The MGnify contig viewer allows visualization of functional annotation of contigs,
https://www.ebi.ac.uk/metagenomics/analyses/MGYA00383254#contigs-viewer
. In this particular view, the contig ERZ477576.854-NODE-854-length-9986-cov-4.0004:1-9986 has been selected and the proteins coloured according to the COG category, with the four cytochrome c subunits highlighted.
Similar articles
- MGnify: the microbiome sequence data analysis resource in 2023.
Richardson L, Allen B, Baldi G, Beracochea M, Bileschi ML, Burdett T, Burgin J, Caballero-Pérez J, Cochrane G, Colwell LJ, Curtis T, Escobar-Zepeda A, Gurbich TA, Kale V, Korobeynikov A, Raj S, Rogers AB, Sakharova E, Sanchez S, Wilkinson DJ, Finn RD. Richardson L, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D753-D759. doi: 10.1093/nar/gkac1080. Nucleic Acids Res. 2023. PMID: 36477304 Free PMC article. - EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies.
Mitchell AL, Scheremetjew M, Denise H, Potter S, Tarkowska A, Qureshi M, Salazar GA, Pesseat S, Boland MA, Hunter FMI, Ten Hoopen P, Alako B, Amid C, Wilkinson DJ, Curtis TP, Cochrane G, Finn RD. Mitchell AL, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D726-D735. doi: 10.1093/nar/gkx967. Nucleic Acids Res. 2018. PMID: 29069476 Free PMC article. - MGnify Genomes: A Resource for Biome-specific Microbial Genome Catalogues.
Gurbich TA, Almeida A, Beracochea M, Burdett T, Burgin J, Cochrane G, Raj S, Richardson L, Rogers AB, Sakharova E, Salazar GA, Finn RD. Gurbich TA, et al. J Mol Biol. 2023 Jul 15;435(14):168016. doi: 10.1016/j.jmb.2023.168016. Epub 2023 Feb 16. J Mol Biol. 2023. PMID: 36806692 Free PMC article. - Benchmarking Metagenomics Tools for Taxonomic Classification.
Ye SH, Siddle KJ, Park DJ, Sabeti PC. Ye SH, et al. Cell. 2019 Aug 8;178(4):779-794. doi: 10.1016/j.cell.2019.07.010. Cell. 2019. PMID: 31398336 Free PMC article. Review. - Microbial diversity of hypersaline environments: a metagenomic approach.
Ventosa A, de la Haba RR, Sánchez-Porro C, Papke RT. Ventosa A, et al. Curr Opin Microbiol. 2015 Jun;25:80-7. doi: 10.1016/j.mib.2015.05.002. Epub 2015 Jun 8. Curr Opin Microbiol. 2015. PMID: 26056770 Review.
Cited by
- The Innovative Informatics Approaches of High-Throughput Technologies in Livestock: Spearheading the Sustainability and Resiliency of Agrigenomics Research.
Suminda GGD, Ghosh M, Son YO. Suminda GGD, et al. Life (Basel). 2022 Nov 15;12(11):1893. doi: 10.3390/life12111893. Life (Basel). 2022. PMID: 36431028 Free PMC article. Review. - TCR signaling induces STAT3 phosphorylation to promote TH17 cell differentiation.
Qin Z, Wang R, Hou P, Zhang Y, Yuan Q, Wang Y, Yang Y, Xu T. Qin Z, et al. J Exp Med. 2024 Mar 4;221(3):e20230683. doi: 10.1084/jem.20230683. Epub 2024 Feb 7. J Exp Med. 2024. PMID: 38324068 Free PMC article. - Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search.
Bryant P, Pozzati G, Zhu W, Shenoy A, Kundrotas P, Elofsson A. Bryant P, et al. Nat Commun. 2022 Oct 12;13(1):6028. doi: 10.1038/s41467-022-33729-4. Nat Commun. 2022. PMID: 36224222 Free PMC article. - Microbiome and oral squamous cell carcinoma: a possible interplay on iron metabolism and its impact on tumor microenvironment.
Arthur RA, Dos Santos Bezerra R, Ximenez JPB, Merlin BL, de Andrade Morraye R, Neto JV, Fava NMN, Figueiredo DLA, de Biagi CAO Jr; Summer Course 2020 group; Montibeller MJ, Guimarães JB, Alves EG, Schreiner M, da Costa TS, da Silva CFL, Malheiros JM, da Silva LHB, Ribas GT, Achallma DO, Braga CM, Andrade KFA, do Carmo Alves Martins V, Dos Santos GVN, Granatto CF, Terin UC, Sanches IH, Ramos DE, Garay-Malpartida HM, de Souza GMP, Slavov SN, Silva WA Jr. Arthur RA, et al. Braz J Microbiol. 2021 Sep;52(3):1287-1302. doi: 10.1007/s42770-021-00491-6. Epub 2021 May 18. Braz J Microbiol. 2021. PMID: 34002353 Free PMC article. - Distance-AF: Modifying Predicted Protein Structure Models by Alphafold2 with User-Specified Distance Constraints.
Zhang Y, Zhang Z, Kagaya Y, Terashi G, Zhao B, Xiong Y, Kihara D. Zhang Y, et al. bioRxiv [Preprint]. 2023 Dec 4:2023.12.01.569498. doi: 10.1101/2023.12.01.569498. bioRxiv. 2023. PMID: 38106200 Free PMC article. Preprint.
References
- Overholt W.A., Schwing P., Raz K.M., Hastings D., Hollander D.J., Kostka J.E.. The core seafloor microbiome in the Gulf of Mexico is remarkably consistent and shows evidence of recovery from disturbance caused by major oil spills. Environ. Microbiol. 2019; doi:10.1111/1462-2920.14794. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/M011755/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R015228/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N018354/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous