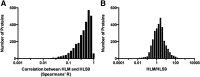Comparative Proteomics Analysis of Human Liver Microsomes and S9 Fractions - PubMed (original) (raw)
Comparative Study
Comparative Proteomics Analysis of Human Liver Microsomes and S9 Fractions
Xinwen Wang et al. Drug Metab Dispos. 2020 Jan.
Abstract
Human liver microsomes (HLM) and human liver S9 fractions (HLS9) are commonly used to study drug metabolism in vitro. However, a quantitative comparison of HLM and HLS9 proteomes is lacking, resulting in the arbitrary selection of one hepatic preparation over another and in difficulties with data interpretation. In this study, we applied a label-free global absolute quantitative proteomics method to the analysis of HLS9 and the corresponding HLM prepared from 102 individual human livers. A total of 3137 proteins were absolutely quantified, and 3087 of those were determined in both HLM and HLS9. Protein concentrations were highly correlated between the two hepatic preparations (R = 0.87, P < 0.0001). We reported the concentrations of 98 drug-metabolizing enzymes (DMEs) and 51 transporters, and demonstrated significant differences between their abundances in HLM and HLS9. We also revealed the protein-protein correlations among these DMEs and transporters and the sex effect on the HLM and HLS9 proteomes. Additionally, HLM and HLS9 displayed distinct expression patterns for protein markers of cytosol and various cellular organelles. Moreover, we evaluated the interindividual variability of three housekeeping proteins, and identified five proteins with low variation across individuals that have the potential to serve as new internal controls for western blot experiments. In summary, these results will lead to better understanding of data obtained from HLM and HLS9 and assist in in vitro-in vivo extrapolations. Knowing the differences between HLM and HLS9 also allows us to make better-informed decisions when choosing between these two hepatic preparations for in vitro drug metabolism studies. SIGNIFICANCE STATEMENT: This investigation revealed significant differences in protein concentrations of drug-metabolizing enzymes and transporters between human liver microsomes and S9 fractions. We also determined the protein-protein correlations among the drug-metabolizing enzymes and transporters and the sex effect on the proteomes of these two hepatic preparations. The results will help interpret data obtained from these two preparations and allow us to make more informed decisions when choosing between human liver microsomes and S9 fractions for in vitro drug metabolism studies.
Copyright © 2019 by The American Society for Pharmacology and Experimental Therapeutics.
Figures
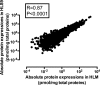
Fig. 1.
Correlation between the mean protein abundances in the 102 HLM and the matched HLS9 samples. Absolute protein concentrations were obtained using the label-free DIA-TPA method. Spearman’s correlation analysis was performed and P < 0.05 was considered statistically significant.
Fig. 2.
Distributions of the Spearman’s R values of the protein concentration correlations between HLM and HLS9 (A) and the HLM-to-HLS9 ratios of mean protein concentrations (B). Fifty proteins that were only quantified in one type of samples were removed from the analysis.
Fig. 3.
Mean protein concentrations of P450s (A), hydrolases (B), UGTs (C), and other transferases (D) in the 102 matched individual HLM and HLS9 samples. Black and gray bars represent protein concentrations in HLM and HLS9, respectively, and error bars represent S.D.
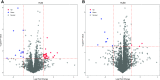
Fig. 4.
The differences in the liver proteomes between males and females in HLM (A) and HLS9 (B). The _x_-axis is the log2 protein concentration ratios of males to females, with the two vertical dotted lines indicating a 2-fold difference between males and females. The _y_-axis indicates the −log10 (P value) with a horizontal dotted line at P = 0.05, and the higher the value of y, the more significant is the difference. The red and blue dots represent the proteins being significantly higher in males and females, respectively.
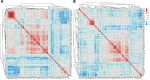
Fig. 5.
Heat map of protein-protein correlation analysis in HLM (A) and HLS9 (B): red- and blue-shaded boxes indicate the extent of positive and negative correlations, respectively; white boxes indicate no correlation.
Similar articles
- Global Proteomic Analysis of Human Liver Microsomes: Rapid Characterization and Quantification of Hepatic Drug-Metabolizing Enzymes.
Achour B, Al Feteisi H, Lanucara F, Rostami-Hodjegan A, Barber J. Achour B, et al. Drug Metab Dispos. 2017 Jun;45(6):666-675. doi: 10.1124/dmd.116.074732. Epub 2017 Apr 3. Drug Metab Dispos. 2017. PMID: 28373266 - A Comparative Analysis of Cytochrome P450 Activities in Paired Liver and Small Intestinal Samples from Patients with Obesity.
Krogstad V, Peric A, Robertsen I, Kringen MK, Wegler C, Angeles PC, Hjelmesæth J, Karlsson C, Andersson S, Artursson P, Åsberg A, Andersson TB, Christensen H. Krogstad V, et al. Drug Metab Dispos. 2020 Jan;48(1):8-17. doi: 10.1124/dmd.119.087940. Epub 2019 Nov 4. Drug Metab Dispos. 2020. PMID: 31685482 - Differential Tissue Abundance of Membrane-Bound Drug Metabolizing Enzymes and Transporter Proteins by Global Proteomics.
Singh DK, Ahire D, Davydov DR, Prasad B. Singh DK, et al. Drug Metab Dispos. 2024 Oct 16;52(11):1152-1160. doi: 10.1124/dmd.124.001477. Drug Metab Dispos. 2024. PMID: 38641346 - Platelet-rich therapies for musculoskeletal soft tissue injuries.
Moraes VY, Lenza M, Tamaoki MJ, Faloppa F, Belloti JC. Moraes VY, et al. Cochrane Database Syst Rev. 2014 Apr 29;2014(4):CD010071. doi: 10.1002/14651858.CD010071.pub3. Cochrane Database Syst Rev. 2014. PMID: 24782334 Free PMC article. Review. - Nutritional supplementation for stable chronic obstructive pulmonary disease.
Ferreira IM, Brooks D, White J, Goldstein R. Ferreira IM, et al. Cochrane Database Syst Rev. 2012 Dec 12;12(12):CD000998. doi: 10.1002/14651858.CD000998.pub3. Cochrane Database Syst Rev. 2012. PMID: 23235577 Free PMC article. Review.
Cited by
- Nonadditivity in human microsomal drug metabolism revealed in a study with coumarin 152, a polyspecific cytochrome P450 substrate.
Dangi B, Davydova NY, Vavilov NE, Zgoda VG, Davydov DR. Dangi B, et al. Xenobiotica. 2020 Dec;50(12):1393-1405. doi: 10.1080/00498254.2020.1775913. Epub 2020 Jul 26. Xenobiotica. 2020. PMID: 32662751 Free PMC article. - Fluorogenic chemical tools to shed light on CES1-mediated adverse drug interactions.
Karns CJ, Spidle TP, Adusah E, Gao M, Nehls JE, Beck MW. Karns CJ, et al. Chem Commun (Camb). 2024 Oct 22;60(85):12369-12372. doi: 10.1039/d4cc03632h. Chem Commun (Camb). 2024. PMID: 39279555 - Genome-Wide Association Study for the Genetic Determinants of Thiopurine Methyltransferase Protein Expression in Human Livers and Racial Differences.
Smith LS, Wang X, Shi J, He B, Zhu HJ. Smith LS, et al. Pharm Res. 2023 Nov;40(11):2525-2531. doi: 10.1007/s11095-023-03558-1. Epub 2023 Jul 10. Pharm Res. 2023. PMID: 37430149 - Novel Independent Trans- and Cis-Genetic Variants Associated with CYP2D6 Expression and Activity in Human Livers.
Smith D, He B, Shi J, Zhu HJ, Wang X. Smith D, et al. Drug Metab Dispos. 2024 Jan 9;52(2):143-152. doi: 10.1124/dmd.123.001548. Drug Metab Dispos. 2024. PMID: 38050015 Free PMC article. - Developmental Expression of Drug Transporters and Conjugating Enzymes Involved in Enterohepatic Recycling: Implication for Pediatric Drug Dosing.
Thakur A, Subash S, Ahire D, Prasad B. Thakur A, et al. Clin Pharmacol Ther. 2024 Dec;116(6):1615-1626. doi: 10.1002/cpt.3409. Epub 2024 Aug 19. Clin Pharmacol Ther. 2024. PMID: 39160670
References
- Achour B, Al Feteisi H, Lanucara F, Rostami-Hodjegan A, Barber J. (2017a) Global proteomic analysis of human liver microsomes: rapid characterization and quantification of hepatic drug-metabolizing enzymes. Drug Metab Dispos 45:666–675. - PubMed
- Achour B, Barber J, Rostami-Hodjegan A. (2014a) Expression of hepatic drug-metabolizing cytochrome P450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 42:1349–1356. - PubMed
- Achour B, Dantonio A, Niosi M, Novak JJ, Fallon JK, Barber J, Smith PC, Rostami-Hodjegan A, Goosen TC. (2017b) Quantitative characterization of major hepatic UDP-glucuronosyltransferase enzymes in human liver microsomes: comparison of two proteomic methods and correlation with catalytic activity. Drug Metab Dispos 45:1102–1112. - PubMed
- Achour B, Russell MR, Barber J, Rostami-Hodjegan A. (2014b) Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos 42:500–510. - PubMed
- Asha S, Vidyavathi M. (2010) Role of human liver microsomes in in vitro metabolism of drugs—a review. Appl Biochem Biotechnol 160:1699–1722. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical