Conserved Patterns of Symmetric Inversion in the Genome Evolution of Bordetella Respiratory Pathogens - PubMed (original) (raw)
. 2019 Nov 19;4(6):e00702-19.
doi: 10.1128/mSystems.00702-19.
Yanhui Peng 2, Dhwani Batra 3, Mark Burroughs 3, Jamie K Davis 3, Kristen Knipe 3, Vladimir N Loparev 3, Taccara Johnson 2, Phalasy Juieng 3, Lori A Rowe 3, Mili Sheth 3, Kevin Tang 3, Yvette Unoarumhi 3, Margaret M Williams 2, M Lucia Tondella 2
Affiliations
- PMID: 31744907
- PMCID: PMC6867878
- DOI: 10.1128/mSystems.00702-19
Conserved Patterns of Symmetric Inversion in the Genome Evolution of Bordetella Respiratory Pathogens
Michael R Weigand et al. mSystems. 2019.
Abstract
Whooping cough (pertussis), primarily caused by Bordetella pertussis, has resurged in the United States, and circulating strains exhibit considerable chromosome structural fluidity in the form of rearrangement and deletion. The genus Bordetella includes additional pathogenic species infecting various animals, some even causing pertussis-like respiratory disease in humans; however, investigation of their genome evolution has been limited. We studied chromosome structure in complete genome sequences from 167 Bordetella species isolates, as well as 469 B. pertussis isolates, to gain a generalized understanding of rearrangement patterns among these related pathogens. Observed changes in gene order primarily resulted from large inversions and were only detected in species with genomes harboring multicopy insertion sequence (IS) elements, most notably B. holmesii and B. parapertussis While genomes of B. pertussis contain >240 copies of IS_481_, IS elements appear less numerous in other species and yield less chromosome structural diversity through rearrangement. These data were further used to predict all possible rearrangements between IS element copies present in Bordetella genomes, revealing that only a subset is observed among circulating strains. Therefore, while it appears that rearrangement occurs less frequently in other species than in B. pertussis, these clinically relevant respiratory pathogens likely experience similar mutation of gene order. The resulting chromosome structural fluidity presents both challenges and opportunity for the study of Bordetella respiratory pathogens.IMPORTANCE Bordetella pertussis is the primary agent of whooping cough (pertussis). The Bordetella genus includes additional pathogens of animals and humans, including some that cause pertussis-like respiratory illness. The chromosome of B. pertussis has previously been shown to exhibit considerable structural rearrangement, but insufficient data have prevented comparable investigation in related species. In this study, we analyze chromosome structure variation in several Bordetella species to gain a generalized understanding of rearrangement patterns in this genus. Just as in B. pertussis, we observed inversions in other species that likely result from common mutational processes. We used these data to further predict additional, unobserved inversions, suggesting that specific genome structures may be preferred in each species.
Keywords: Bordetella; evolution; genomics; insertion sequence; pertussis; rearrangement; whooping cough.
Figures
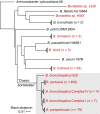
FIG 1
Phylogeny of the genus Bordetella. Neighbor-joining tree of named and provisional Bordetella species with available complete-genome sequences based on pairwise mash distances and rooted with Achromobacter xylosoxidans. Species sequenced at the CDC are listed in red. Triangles indicate collapsed nodes.
FIG 2
Genome rearrangements in Bordetella species. (A) Eleven unique chromosome structures that included six symmetric inversions were observed among B. holmesii isolate genomes. (B) Two predominant structures were observed in B. parapertussis. (C, D) Phylogenetic placement of structures observed in B. holmesii (C) and B. parapertussis (D) isolates was reconstructed using maximum parsimony from 1,496 and 677 core SNPs, respectively.
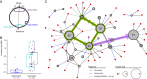
FIG 3
Genome rearrangement in B. pertussis. (A) Observed inversions were either symmetric, encompassing either replication origin (purple) or terminus (light blue), or asymmetric (blue). (B) Single inversions observed in pairwise alignments varied in size and were predominantly symmetric. (C) Undirected network constructed from single inversions or insertion/deletions observed between 71 unique chromosome structures. Node diameter and edge line type indicate cluster size and rearrangement, respectively, according to the key. Nodes with the highest degrees of centrality (Fig. S5A) and their connections at the network core are highlighted (green). Shaded arrows indicate directionality inferred from the SNP phylogeny (Fig. S7), including divergence toward the predominant cluster BP-01 (pink), as described in the text.
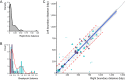
FIG 4
Symmetric inversion balance and prediction. (A) Density of relative replichore size balance observed in each of the 107 unique B. pertussis genome structures. (B) Histogram of breakpoint balance observed in symmetric inversions encompassing the replication origin (purple) or terminus (light blue). The combined densities of observed (solid line) and predicted (dotted line) symmetric inversions were scaled to 10. (C) Linear regression of right and left breakpoint distances observed in B. pertussis (circle) and B. holmesii (triangle) nearest to the replication origin (purple) or terminus (light blue). Open symbols represent duplicated points inverted for model calculation. Blue line indicates linear fit, and shading represents the 95% confidence interval. Dotted red lines denote boundaries of the model’s 95% prediction interval, and gray points correspond to all predicted inversions between copies of IS elements in B. pertussis J549.
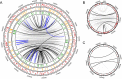
FIG 5
Inversion potential of Bordetella species genomes based on IS element content. Chromosome maps of B. pertussis J549 (A), B. holmesii C690 (B), and B. parapertussis B271 (C). Tracks of red lines indicate the locations of IS elements in either the forward or reverse orientation (above or below center, respectively). Connecting lines link breakpoints of observed symmetric (black), observed asymmetric (blue), and predicted symmetric (gray) inversions. Additional tracks in panel A denote all breakpoints observed in pairwise alignments between J549 and genomes of the ptxP1-ptxP2 (orange) or ptxP3 (green) clades.
Similar articles
- The History of Bordetella pertussis Genome Evolution Includes Structural Rearrangement.
Weigand MR, Peng Y, Loparev V, Batra D, Bowden KE, Burroughs M, Cassiday PK, Davis JK, Johnson T, Juieng P, Knipe K, Mathis MH, Pruitt AM, Rowe L, Sheth M, Tondella ML, Williams MM. Weigand MR, et al. J Bacteriol. 2017 Mar 28;199(8):e00806-16. doi: 10.1128/JB.00806-16. Print 2017 Apr 15. J Bacteriol. 2017. PMID: 28167525 Free PMC article. - Insertion sequences shared by Bordetella species and implications for the biological diagnosis of pertussis syndrome.
Tizolova A, Guiso N, Guillot S. Tizolova A, et al. Eur J Clin Microbiol Infect Dis. 2013 Jan;32(1):89-96. doi: 10.1007/s10096-012-1718-3. Epub 2012 Aug 12. Eur J Clin Microbiol Infect Dis. 2013. PMID: 22886091 - Genome Structural Diversity among 31 Bordetella pertussis Isolates from Two Recent U.S. Whooping Cough Statewide Epidemics.
Bowden KE, Weigand MR, Peng Y, Cassiday PK, Sammons S, Knipe K, Rowe LA, Loparev V, Sheth M, Weening K, Tondella ML, Williams MM. Bowden KE, et al. mSphere. 2016 May 11;1(3):e00036-16. doi: 10.1128/mSphere.00036-16. eCollection 2016 May-Jun. mSphere. 2016. PMID: 27303739 Free PMC article. - Other Bordetellas, lessons for and from pertussis vaccines.
Guiso N, Hegerle N. Guiso N, et al. Expert Rev Vaccines. 2014 Sep;13(9):1125-33. doi: 10.1586/14760584.2014.942221. Epub 2014 Jul 18. Expert Rev Vaccines. 2014. PMID: 25034039 Review. - Genomics of Bordetella pertussis toxins.
Antoine R, Raze D, Locht C. Antoine R, et al. Int J Med Microbiol. 2000 Oct;290(4-5):301-5. doi: 10.1016/S1438-4221(00)80026-0. Int J Med Microbiol. 2000. PMID: 11111902 Review.
Cited by
- Replication-associated inversions are the dominant form of bacterial chromosome structural variation.
D'Iorio M, Dewar K. D'Iorio M, et al. Life Sci Alliance. 2022 Oct 19;6(1):e202201434. doi: 10.26508/lsa.202201434. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36261227 Free PMC article. - A 69.9-kb long inverted repeat increases genome instability in a strain of Lactobacillus crispatus.
Colombini L, Santoro F, Tirziu M, Cuppone AM, Pozzi G, Iannelli F. Colombini L, et al. NAR Genom Bioinform. 2025 Jun 26;7(2):lqaf085. doi: 10.1093/nargab/lqaf085. eCollection 2025 Jun. NAR Genom Bioinform. 2025. PMID: 40575252 Free PMC article. - Tatajuba: exploring the distribution of homopolymer tracts.
de Oliveira Martins L, Bloomfield S, Stoakes E, Grant AJ, Page AJ, Mather AE. de Oliveira Martins L, et al. NAR Genom Bioinform. 2022 Feb 2;4(1):lqac003. doi: 10.1093/nargab/lqac003. eCollection 2022 Mar. NAR Genom Bioinform. 2022. PMID: 35118377 Free PMC article. - Bordetella spp. block eosinophil recruitment to suppress the generation of early mucosal protection.
First NJ, Parrish KM, Martínez-Pérez A, González-Fernández Á, Bharrhan S, Woolard M, McLachlan JB, Scott RS, Wang J, Gestal MC. First NJ, et al. Cell Rep. 2023 Nov 28;42(11):113294. doi: 10.1016/j.celrep.2023.113294. Epub 2023 Oct 25. Cell Rep. 2023. PMID: 37883230 Free PMC article. - Cytochrome oxidase requirements in Bordetella reveal insights into evolution towards life in the mammalian respiratory tract.
McKay LS, Spandrio AR, Johnson RM, Sobran MA, Marlatt SA, Mote KB, Dedloff MR, Nash ZM, Julio SM, Cotter PA. McKay LS, et al. PLoS Pathog. 2024 Jul 8;20(7):e1012084. doi: 10.1371/journal.ppat.1012084. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 38976749 Free PMC article.
References
- von Wintzingerode F, Gerlach G, Schneider B, Gross R. 2002. Phylogenetic relationships and virulence evolution in the genus Bordetella. Curr Top Microbiol Immunol 264:177–199. - PubMed
- Park J, Zhang Y, Buboltz AM, Zhang X, Schuster SC, Ahuja U, Liu M, Miller JF, Sebaihia M, Bentley SD, Parkhill J, Harvill ET. 2012. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13:545. doi:10.1186/1471-2164-13-545. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases