Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases - PubMed (original) (raw)
Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases
Daniela Šimčíková et al. Sci Rep. 2019.
Abstract
Prediction methods have become an integral part of biomedical and biotechnological research. However, their clinical interpretations are largely based on biochemical or molecular data, but not clinical data. Here, we focus on improving the reliability and clinical applicability of prediction algorithms. We assembled and curated two large non-overlapping large databases of clinical phenotypes. These phenotypes were caused by missense variations in 44 and 63 genes associated with Mendelian diseases. We used these databases to establish and validate the model, allowing us to improve the predictions obtained from EVmutation, SNAP2 and PoPMuSiC 2.1. The predictions of clinical effects suffered from a lack of specificity, which appears to be the common constraint of all recently used prediction methods, although predictions mediated by these methods are associated with nearly absolute sensitivity. We introduced evidence-based tailoring of the default settings of the prediction methods; this tailoring substantially improved the prediction outcomes. Additionally, the comparisons of the clinically observed and theoretical variations led to the identification of large previously unreported pools of variations that were under negative selection during molecular evolution. The evolutionary variation analysis approach described here is the first to enable the highly specific identification of likely disease-causing missense variations that have not yet been associated with any clinical phenotype.
Conflict of interest statement
D.S. and P.H. have been funded by the Czech Science Foundation project 15-03834Y and Charles University in Prague projects Primus/MED/32, GA UK 1428218 and 260387/SVV/2017. The authors declare no other competing interests.
Figures
Figure 1
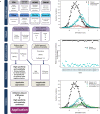
The efficiency of the EVmutation method in predicting the effects of missense variations with known clinical Mendelian disease-associated phenotypes. (a) Flowchart showing the sources and approaches used for data retrieval, the construction of datasets and subsequent analyses. The selection of analyzed genes associated with Mendelian diseases was based on combined information retrieved from the Human Gene Mutation Database (HGMD), UniProtKB/Swiss-Prot, Protein Data Bank (PDB) and Online Mendelian Inheritance in Man (OMIM). Information about disease associations and no-phenotype associations of clinically observed variations was retrieved from the ClinVar database and the Ensembl browser. Additional information about proteins (domains) and variations (frequency) was obtained from the Pfam database and the Exome Aggregation Consortium (ExAC) browser, respectively. A vertical line indicates the arbitrary threshold for variations with an effect. (b) The distribution of numerical EVmutation scores calculated for missense variations with known clinical phenotypes. (c) The relative percentage of correct predictions of disease and no clinical phenotypes using EVmutation scores calculated for the 44 analyzed proteins. (d) The distribution of numerical EVmutation scores calculated for disease-associated and no phenotype-associated missense variations with known clinical phenotypes in 44 proteins that cause Mendelian diseases sorted according to the evolutionary conservation of affected amino acids in mammals. Conserved amino acids (GV = 0) were conserved in all ten examined mammalian orthologs. Variable amino acids (GV > 0) were not conserved in at least one of the ten examined mammalian orthologs of the respective protein.
Figure 2
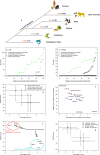
The predictions differ for evolutionarily conserved proteins, such as AR or PTEN, for variations within and outside of protein domains and for enzymes and proteins without enzymatic functions. (a) Evolutionary divergence of the amino acid sequences of AR and PTEN reported as the number of amino acid substitutions per site by averaging all sequence pairs between primates and other groups. (b,c) GV scores for amino acids within the AR (b) and PTEN (c) sequences. The data are shown separately for GV scores calculated based on mammalian protein orthologs (the two lines at the zero GV score) and extended MSAs that included more evolutionarily distant taxa. The data are shown for disease-associated and no phenotype-associated variations. Relative ranks among tested variations are shown to reflect the different numbers of variations included in each analyzed group. (d) EVmutation and SNAP2 scores applied to disease-associated and no phenotype-associated variations that are present or absent from protein domains. Data are presented as medians ± SD. (e) Differences in median EVmutation and SNAP2 scores between disease-associated and no phenotype-associated variations located within the indicated protein domains. Abbreviations for the domains: AGAL, alpha-galactosidase A; ATCase/OTCase, aspartate/ornithine carbamoyltransferase, carbamoyl-P binding and Asp/Orn binding domains; CPOX, coproporphyrinogen III oxidase; DHE1, dehydrogenase E1 component; FRNADBD, ferric reductase, NAD binding domain; GTPCH, GTP cyclohydrolase I; G6PDH, glucose-6-phosphate dehydrogenase, NAD binding and C-terminal domains; HXK, hexokinase; LBDNHR, ligand-binding domain of nuclear hormone receptor; PK, protein kinase; PTK, protein tyrosine kinase; PTP SH2, Src Homology 2 domain. (f) Median EVmutation and SNAP2 scores calculated for disease-associated and no phenotype-associated variations in the four indicated enzyme classes and in proteins without enzymatic functions. (g) EVmutation and SNAP2 scores calculated for disease-associated and no phenotype-associated variations considered possible or impossible variations according to Bromberg et al. Data are shown as medians ± SD.
Figure 3
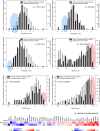
The efficiency of the prediction methods in discriminating among multiple diseases caused by missense variations in the indicated proteins. EVmutation and SNAP2 scores are shown for proteins with significantly different disease-specific scores (a–i) or that result in the opposite phenotypes (j–l). (a) DMD, (b) ELANE, (c) FLNA, (d) HPRT1, (e) PTPN11, (f) RET, (g) TGFRB2, (h) TP63, (i) UROD, (j) GCK, (k) HNF4A, and (l) HBB.
Figure 4
The detection of variations under negative selection during molecular evolution: an example of the application of evidence-based knowledge. (a–f) The distribution of observed disease-associated variations compared to the distribution of possible theoretical variations. The data are shown for the proteins for which negative values were obtained from the calculation of the differences in the 10th percentiles of EVmutation scores – (a) PTPN11, (b) HBB and (c) G6PD – and for genes for which positive values were obtained from the calculation of the differences in 90th percentiles of SNAP2 scores – (d) G6PD, (e) HNF4A and (f) EDA. (g) The heatmap of proteins causing Mendelian diseases sorted according to the likelihood that their variations included variations that were under negative selection during molecular evolution. Ranges of differences in median values: from −1.093 to 3.36 (EVmutation) and from −25 to 2.6 (SNAP2).
Figure 5
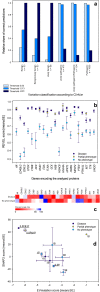
Validation of the model, identification of the specificity of the consensus classifier REVEL, and the application of the American College of Medical Genetics and Genomics (ACMG) criteria for the classification of variations. (a) Validation of the threshold values for EVmutation that were suggested in the proposed model. Validation was performed using a set of 1723 variations in 63 genes (Tables S8–S10), which were classified according to ClinVar. The data are presented as relative percentages of correct predictions using the arbitrary EVmutation threshold (0.00), the evidence-based threshold that allows 95% sensitivity (−2.13) and the threshold that allows 95% specificity (−8.81). (b,c) REVEL, a consensus classifier, is associated with the issue of low specificity, similar to the individual computational algorithms. REVEL scores were retrieved for a set of 2721 variations in 21 genes. Mean REVEL scores for the individual genes discriminated well between the disease-associated and no phenotype-associated variations (b). However, because a large overlap in the predictions was observed, the specificity was low for most of the analyzed genes (c). Data are presented (b) as the means ± SE or (c) as relative percentages of correct predictions of the association of the variations with diseases (upper row) or no phenotypes (lower row). (d) Application of the ACMG criteria for the classification of variations, which classify the variations as benign (1B and higher) and pathogenic (0.5 P and higher) according to the population frequencies of the variations (Table S11). The EVmutation and SNAP2 scores were analyzed separately for the disease- and no phenotype-associated variations. Data are shown as means ± SE.
Similar articles
- Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information.
Capriotti E, Calabrese R, Casadio R. Capriotti E, et al. Bioinformatics. 2006 Nov 15;22(22):2729-34. doi: 10.1093/bioinformatics/btl423. Epub 2006 Aug 7. Bioinformatics. 2006. PMID: 16895930 - Evidence-based tailoring of bioinformatics approaches to optimize methods that predict the effects of nonsynonymous amino acid substitutions in glucokinase.
Šimčíková D, Kocková L, Vackářová K, Těšínský M, Heneberg P. Šimčíková D, et al. Sci Rep. 2017 Aug 25;7(1):9499. doi: 10.1038/s41598-017-09810-0. Sci Rep. 2017. PMID: 28842611 Free PMC article. - In silico analysis of missense mutations in exons 1-5 of the F9 gene that cause hemophilia B.
Meléndez-Aranda L, Jaloma-Cruz AR, Pastor N, Romero-Prado MMJ. Meléndez-Aranda L, et al. BMC Bioinformatics. 2019 Jun 28;20(1):363. doi: 10.1186/s12859-019-2919-x. BMC Bioinformatics. 2019. PMID: 31253089 Free PMC article. - Objective assessment of the evolutionary action equation for the fitness effect of missense mutations across CAGI-blinded contests.
Katsonis P, Lichtarge O. Katsonis P, et al. Hum Mutat. 2017 Sep;38(9):1072-1084. doi: 10.1002/humu.23266. Epub 2017 Jun 21. Hum Mutat. 2017. PMID: 28544059 Free PMC article. Review. - Disease gene discovery through integrative genomics.
Giallourakis C, Henson C, Reich M, Xie X, Mootha VK. Giallourakis C, et al. Annu Rev Genomics Hum Genet. 2005;6:381-406. doi: 10.1146/annurev.genom.6.080604.162234. Annu Rev Genomics Hum Genet. 2005. PMID: 16124867 Review.
Cited by
- Exploring TTN variants as genetic insights into cardiomyopathy pathogenesis and potential emerging clues to molecular mechanisms in cardiomyopathies.
Jolfayi AG, Kohansal E, Ghasemi S, Naderi N, Hesami M, MozafaryBazargany M, Moghadam MH, Fazelifar AF, Maleki M, Kalayinia S. Jolfayi AG, et al. Sci Rep. 2024 Mar 4;14(1):5313. doi: 10.1038/s41598-024-56154-7. Sci Rep. 2024. PMID: 38438525 Free PMC article. Review. - Apolipoprotein E (APOE) Haplotypes in Healthy Subjects from Worldwide Macroareas: A Population Genetics Perspective for Cardiovascular Disease, Neurodegeneration, and Dementia.
Abondio P, Bruno F, Luiselli D. Abondio P, et al. Curr Issues Mol Biol. 2023 Mar 31;45(4):2817-2831. doi: 10.3390/cimb45040184. Curr Issues Mol Biol. 2023. PMID: 37185708 Free PMC article. - Mapping the Constrained Coding Regions in the Human Genome to Their Corresponding Proteins.
Hasenahuer MA, Sanchis-Juan A, Laskowski RA, Baker JA, Stephenson JD, Orengo CA, Raymond FL, Thornton JM. Hasenahuer MA, et al. J Mol Biol. 2023 Jan 30;435(2):167892. doi: 10.1016/j.jmb.2022.167892. Epub 2022 Nov 21. J Mol Biol. 2023. PMID: 36410474 Free PMC article. - The missing heritability in type 1 diabetes.
Pang H, Lin J, Luo S, Huang G, Li X, Xie Z, Zhou Z. Pang H, et al. Diabetes Obes Metab. 2022 Oct;24(10):1901-1911. doi: 10.1111/dom.14777. Epub 2022 Jun 13. Diabetes Obes Metab. 2022. PMID: 35603907 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous