Phylogenomics provides robust support for a two-domains tree of life - PubMed (original) (raw)
Phylogenomics provides robust support for a two-domains tree of life
Tom A Williams et al. Nat Ecol Evol. 2020 Jan.
Erratum in
- Author Correction: Phylogenomics provides robust support for a two-domains tree of life.
Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM. Williams TA, et al. Nat Ecol Evol. 2020 Nov;4(11):1568. doi: 10.1038/s41559-020-01347-2. Nat Ecol Evol. 2020. PMID: 33077932
Abstract
Hypotheses about the origin of eukaryotic cells are classically framed within the context of a universal 'tree of life' based on conserved core genes. Vigorous ongoing debate about eukaryote origins is based on assertions that the topology of the tree of life depends on the taxa included and the choice and quality of genomic data analysed. Here we have reanalysed the evidence underpinning those claims and apply more data to the question by using supertree and coalescent methods to interrogate >3,000 gene families in archaea and eukaryotes. We find that eukaryotes consistently originate from within the archaea in a two-domains tree when due consideration is given to the fit between model and data. Our analyses support a close relationship between eukaryotes and Asgard archaea and identify the Heimdallarchaeota as the current best candidate for the closest archaeal relatives of the eukaryotic nuclear lineage.
Conflict of interest statement
Competing interests statement: The authors declare they have no competing interests.
Figures
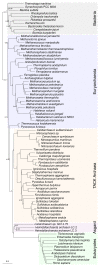
Figure 1. The 35-gene matrix of Da Cunha et al. favours a two-domains tree using the best-fitting models in both maximum likelihood and Bayesian analyses.
The eukaryotes (green) group with the sampled Asgard archaea (orange) with maximum posterior support. Bacteria are in grey, TACK Archaea in yellow, Euryarchaeota in blue. This is a consensus tree inferred under the CAT+GTR+G4 model in PhyloBayes-MPI; branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. A 2D topology was obtained under a variety of other models in ML analyses (LG+G4+F, LG+PMSF+G4, LG+C60+G4+F; Supplementary Figure 1), and also with 4-state Susko-Roger recoding under the CAT+GTR+G4 and NDCH2 models (Supplementary Figure 2).
Figure 2. Evidence that the three-domains tree is an artifact of long branch attraction.
(a) Da Cunha et al. analysed a dataset of 35 core protein-coding genes under the LG+G4+F model and obtained a 3D tree; the better-fitting (Supplementary Table 4) CAT+GTR+G4 model recovers a 2D tree. Bootstrap support (a) and Bayesian posterior probability (b) are indicated for the key nodes defining the 3D and 2D trees. “Asgard” refers to a clade of Heimdallarchaeota and Lokiarchaeum. Plotting these trees to the same scale (in terms of substitutions per site) illustrates major differences in these analyses. The 3D/LG+G4+F analysis suggests that, on average, 30.77 changes have taken place per site; the two-domains/CAT+GTR+G4 analysis suggests that 47.4 changes per site have occurred. This difference amounts to ~128,511 additional substitutions in total inferred under the CAT+GTR+G4 model. (b) Posterior predictive tests indicate that CAT+GTR+G4 performs significantly better than LG+G4+F in capturing the site-specific evolutionary constraints reflected by lower biochemical diversity approaching that of the empirical data). This results in more realistic estimates of substitutional saturation and convergence found in the data. The longest branches on both the 3D and 2D tree are the stems leading to the bacteria and eukaryotes (in blue and green, respectively). CAT+GTR+G4 identifies many more convergent substitutions on these branches than does LG+G4+F, as can be seen by comparing the branch lengths in (a). This failure to detect convergent substitutions under LG+G4+F has the effect of drawing the bacterial and eukaryotic branches together, because convergences are mistaken for homologies (synapomorphies), resulting in a 3D tree.
Figure 3. An expanded sampling of microbial diversity supports a two-domains tree.
(a) Bayesian phylogeny of 21 concatenated proteins conserved across bacteria, archaea and eukaryotes under the CAT+GTR+G4 model, rooted on the branch separating bacteria and archaea. Eukaryotes group with Asgard archaea with maximum posterior support. (b) Bayesian phylogeny of 43 genes conserved between Archaea and eukaryotes under CAT+GTR+G4. Eukaryotes group with, or within, Heimdallarchaeota. All support values are Bayesian posterior probabilities, and branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bars. The Euryarchaeota are paraphyletic in the consensus tree in (a), consistent with some recent analyses using bacterial outgroups,, although the relevant support values are low and the analysis does not robustly exclude the alternative hypothesis of a monophyletic Euryarchaeota. The tree in (b) is formally unrooted because it does not include a bacterial outgroup. Based on (a) and published analyses,, the root may lie between the Euryarchaeota and the other taxa, or within the Euryarchaeota. Amino acid data were recoded using the 4-state scheme of Susko and Roger, which our posterior predictive simulations (Supplementary Table 7) suggest improved model fit by ameliorating substitutional saturation and compositional heterogeneity; phylogenies inferred on the original amino acid data are provided in Supplementary Figure 7.
Comment in
- Evolutionary relationships between archaea and eukaryotes.
Gribaldo S, Brochier-Armanet C. Gribaldo S, et al. Nat Ecol Evol. 2020 Jan;4(1):20-21. doi: 10.1038/s41559-019-1073-1. Nat Ecol Evol. 2020. PMID: 31836857 No abstract available. - Evolution: Two Domains of Life or Three?
Doolittle WF. Doolittle WF. Curr Biol. 2020 Feb 24;30(4):R177-R179. doi: 10.1016/j.cub.2020.01.010. Curr Biol. 2020. PMID: 32097647
Similar articles
- Expanded diversity of Asgard archaea and their relationships with eukaryotes.
Liu Y, Makarova KS, Huang WC, Wolf YI, Nikolskaya AN, Zhang X, Cai M, Zhang CJ, Xu W, Luo Z, Cheng L, Koonin EV, Li M. Liu Y, et al. Nature. 2021 May;593(7860):553-557. doi: 10.1038/s41586-021-03494-3. Epub 2021 Apr 28. Nature. 2021. PMID: 33911286 Free PMC article. - Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes.
Eme L, Tamarit D, Caceres EF, Stairs CW, De Anda V, Schön ME, Seitz KW, Dombrowski N, Lewis WH, Homa F, Saw JH, Lombard J, Nunoura T, Li WJ, Hua ZS, Chen LX, Banfield JF, John ES, Reysenbach AL, Stott MB, Schramm A, Kjeldsen KU, Teske AP, Baker BJ, Ettema TJG. Eme L, et al. Nature. 2023 Jun;618(7967):992-999. doi: 10.1038/s41586-023-06186-2. Epub 2023 Jun 14. Nature. 2023. PMID: 37316666 Free PMC article. - Peculiar morphology of Asgard archaeal cells close to the prokaryote-eukaryote boundary.
Avcı B, Panagiotou K, Albertsen M, Ettema TJG, Schramm A, Kjeldsen KU. Avcı B, et al. mBio. 2025 May 14;16(5):e0032725. doi: 10.1128/mbio.00327-25. Epub 2025 Apr 16. mBio. 2025. PMID: 40237460 Free PMC article. - Two or three domains: a new view of tree of life in the genomics era.
Zhou Z, Liu Y, Li M, Gu JD. Zhou Z, et al. Appl Microbiol Biotechnol. 2018 Apr;102(7):3049-3058. doi: 10.1007/s00253-018-8831-x. Epub 2018 Feb 27. Appl Microbiol Biotechnol. 2018. PMID: 29484479 Review. - An archaeal origin of eukaryotes supports only two primary domains of life.
Williams TA, Foster PG, Cox CJ, Embley TM. Williams TA, et al. Nature. 2013 Dec 12;504(7479):231-6. doi: 10.1038/nature12779. Nature. 2013. PMID: 24336283 Review.
Cited by
- Timing the evolution of phosphorus-cycling enzymes through geological time using phylogenomics.
Boden JS, Zhong J, Anderson RE, Stüeken EE. Boden JS, et al. Nat Commun. 2024 May 2;15(1):3703. doi: 10.1038/s41467-024-47914-0. Nat Commun. 2024. PMID: 38697988 Free PMC article. - Expanded diversity of Asgard archaea and their relationships with eukaryotes.
Liu Y, Makarova KS, Huang WC, Wolf YI, Nikolskaya AN, Zhang X, Cai M, Zhang CJ, Xu W, Luo Z, Cheng L, Koonin EV, Li M. Liu Y, et al. Nature. 2021 May;593(7860):553-557. doi: 10.1038/s41586-021-03494-3. Epub 2021 Apr 28. Nature. 2021. PMID: 33911286 Free PMC article. - Origin of eukaryotes: What can be learned from the first successfully isolated Asgard archaeon.
Albers S, Ashmore J, Pollard T, Spang A, Zhou J. Albers S, et al. Fac Rev. 2022 Jan 27;11:3. doi: 10.12703/r-01-000005. eCollection 2022. Fac Rev. 2022. PMID: 35174363 Free PMC article. - Reconstruction of the rRNA Sequences of LUCA, with Bioinformatic Implication of the Local Similarities Shared by Them.
Men Y, Lu G, Wang Y, Lin J, Xie Q. Men Y, et al. Biology (Basel). 2022 May 29;11(6):837. doi: 10.3390/biology11060837. Biology (Basel). 2022. PMID: 35741358 Free PMC article. - A divide-and-conquer phylogenomic approach based on character supermatrices resolves early steps in the evolution of the Archaea.
Aouad M, Flandrois JP, Jauffrit F, Gouy M, Gribaldo S, Brochier-Armanet C. Aouad M, et al. BMC Ecol Evol. 2022 Jan 5;22(1):1. doi: 10.1186/s12862-021-01952-0. BMC Ecol Evol. 2022. PMID: 34986784 Free PMC article.
References
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. - PubMed
- Roger AJ, Muñoz-Gómez SA, Kamikawa R. The Origin and Diversification of Mitochondria. Curr Biol. 2017;27:R1177–R1192. - PubMed
- Martijn J, Ettema TJG. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–457. - PubMed
- Williams Ta, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources