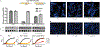A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases - PubMed (original) (raw)
A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases
Senén D Mendoza et al. Nature. 2020 Jan.
Abstract
All viruses require strategies to inhibit or evade the immune pathways of cells that they infect. The viruses that infect bacteria, bacteriophages (phages), must avoid immune pathways that target nucleic acids, such as CRISPR-Cas and restriction-modification systems, to replicate efficiently1. Here we show that jumbo phage ΦKZ segregates its DNA from immunity nucleases of its host, Pseudomonas aeruginosa, by constructing a proteinaceous nucleus-like compartment. ΦKZ is resistant to many immunity mechanisms that target DNA in vivo, including two subtypes of CRISPR-Cas3, Cas9, Cas12a and the restriction enzymes HsdRMS and EcoRI. Cas proteins and restriction enzymes are unable to access the phage DNA throughout the infection, but engineering the relocalization of EcoRI inside the compartment enables targeting of the phage and protection of host cells. Moreover, ΦKZ is sensitive to Cas13a-a CRISPR-Cas enzyme that targets RNA-probably owing to phage mRNA localizing to the cytoplasm. Collectively, we propose that Pseudomonas jumbo phages evade a broad spectrum of DNA-targeting nucleases through the assembly of a protein barrier around their genome.
Conflict of interest statement
Competing Interests: J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences.
Figures
Extended Data Figure 1:. Jumbo phage ΦPA3 resists targeting by CRISPR Cas and a restriction-modification system.
a, Strain PAO1 was engineered to express the I-C cas genes and distinct crRNAs targeting the indicated phages, and plaque assays were conducted as in Fig. 1a. b, Strain PAO1 was engineered to express the Type II-A Cas9 protein and distinct single guide RNAs (sgRNAs) targeting the indicated phage. Plaque assays were conducted as in Figure 1a. c, The endogenous Type I R-M system (hsdRSM) in strain PAO1 was assayed using phages propagated on PAO1 or PAK as indicated (e.g. JBD30•PAO1 was first propagated on strain PAO1). Together with an isogenic PAO1_∆hsdR_ knockout, all strains were subjected to a plaque assay as in Figure 1a. All plaque assays were replicated twice with similar results.
Extended Data Figure 2:. Phage ΦKZ resists P. aeruginosa Type I-C and Type I-F CRISPR-Cas immunity.
a, Strain PAO1 was engineered to express the I-C cas genes and distinct crRNAs targeting phage JBD30 and phage ΦKZ, and plaque assays were conducted as in Fig. 1a. b, Strain PAO1 was engineered to express the I-F cas genes and distinct crRNAs targeting phage JBD30 and phage ΦKZ, and plaque assays were conducted as in Fig. 1a. All plaque assays replicated ≥ 2 times with similar results.
Extended Data Figure 3:. Phage ΦKZ resists targeting by heterologous Type II-A and V-A CRISPR-Cas systems.
a, Strain PAO1 was engineered to express the Type II-A Cas9 protein and distinct single guide RNAs (sgRNAs) targeting the indicated phage. Plaque assays were conducted as in Figure 1a. b, Strain PAO1 was engineered to express the Type V-A Cas12a protein and distinct crRNAs against the indicated phage. Plaque assays were conducted as in Figure 1a. All plaque assays replicated ≥ 2 times with similar results.
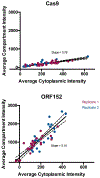
Extended Data Figure 4:. Quantification of Cas9 localization during phage ΦKZ infection of P. aeruginosa.
Localization of Cas9 and ORF152 in the cytoplasm and shell during ΦKZ infection were quantified. Data points (individual cells) from two pooled replicate experiments were fitted with a line, showing 95% confidence intervals with dashed lines. The slope is as reported in the plots.
Extended Data Figure 5:. Phage ΦKZ genomic DNA is susceptible in vitro to cleavage by Restriction Endonucleases.
Genomic DNA was isolated from phages ΦKZ and JBD30 and was subjected to digestion with the indicated restriction enzymes in vitro. The number of cut sites for each enzyme is shown at the bottom of the gels. Products were visualized on a 0.7 % agarose gel, visualized with SYBR Safe nucleic acid stain. In vitro digestion experiment was replicated twice with similar results.
Extended Data Figure 6:. Cas9 fusion to ORF152 localizes to periphery of shell, but does not enable immune activity against ΦKZ.
a, Strain PAO1 was engineered to express Cas9 fused to ORF152 at either the N- or C-terminus, with single guide RNAs (sgRNAs) targeting the indicated phage. Plaque assays were conducted as in Fig. 1a, b, growth curves were conducted, monitoring the OD600 of PAO1 cells infected with the indicated ΦKZ multiplicity of infection (MOI). c, Fluorescence microscopy of PAO1 fusion strains, immunostained for Cas9, in cells expressing ORF152-Cas9 or Cas9-ORF152. DAPI stain shows the phage DNA within the shell. d, Fluorescence microscopy of P. aeruginosa, immunostained for Cas9, in cells expressing ORF152-Cas9. DAPI stain shows the phage DNA within the shell. All plaque assays were replicated 4 times with similar results. Growth curve experiments were replicated three times with similar results. Microscopy was replicated twice with similar results.
Extended Data Figure 7:. Fusion of EcoRI restriction enzymes to ORF152 enables immune activity.
a, Strain PAO1 was engineered to express EcoRI or EcoRI-ORF152 fusion protein. Plaque assays were conducted as in Fig. 1a and quantified (n=3). b, Growth curves were conducted, monitoring the OD600 of PAO1 cells infected with the indicated ΦKZ multiplicity of infection (MOI). c, Live fluorescence imaging of P. aeruginosa strains engineered to express EcoRI or EcoRI-ORF152. DAPI stain shows the phage DNA. d, Live fluorescence imaging of P. aeruginosa strains engineered to express EcoRI, or EcoRI-ORF152. DAPI stain shows the phage DNA within the shell. Wide field of view. All plaque assays were replicated 3 times with similar results. Growth curve experiments were replicated twice with similar results. Microscopy was replicated twice with similar results.
Extended Data Figure 8:. Phage ΦKZ DNA is sensitive to RNA-targeting Cas13.
a, Strain PAO1 expressing LseCas13a and distinct crRNAs targeting the indicated gene. Plaque assays conducted as in Figure 1a. b, Strain PAO1 expressing LshCas13a and distinct crRNAs targeting the indicated gene. Plaque assays conducted as in Figure 1a. All plaque assays were replicated twice with similar results.
Extended Data Figure 9:. Phage ΦKZ escaper mutants are selected by Cas13-mediated RNA targeting.
Strain PAO1 expressing LseCas13a and a crRNA targeting the indicated gene. Plaque assays conducted as in Figure 1a using wild type and escaper mutant ΦKZ. WT ΦKZ is targeted by both strains and the faint clearings observed here are not observed as plaques in a full-plate assay. All plaque assays were replicated twice with similar results.
Extended Data Figure 10:. Observation of DAPI-stained phage DNA adjacent to a Cherry-TopA nascent shell.
Live fluorescence imaging of P. aeruginosa strains engineered to express Cherry-TopA infected with ΦKZ. DAPI stain labels DNA. Microscopy was replicated three times with similar results.
Figure 1:. Identification of a phage that resists targeting by diverse CRISPR-Cas and restriction-modification systems.
Plaque assay with the indicated phage spotted in ten-fold serial diluations on a lawn of P. aeruginosa, dark clearings in the lawn represent phage replication. Strain PAO1 expressing: a, Type I-C cas genes (cas3-5-8-7) and crRNAs targeting the indicated phages. b, Type I-C cas genes and crRNAs targeting phage JBD30 or distinct phage ΦKZ-targeting crRNAs (#1-#3). c, Type II-A cas9 and distinct single guide RNAs (sgRNAs) targeting the indicated phage. d, Type V-A cas12a and distinct crRNAs against the indicated phage. e, Endogenous Type I R-M system (hsdRSM) in strain PAO1 was assayed using phages propagated on PAK (e.g. JBD30•PAK was first propagated on strain PAK). Together with an isogenic PAO1_∆hsdR_ knockout. f, Type II EcoRI R-M system. Restriction activity was assayed using phages JBD30 and ΦKZ. All plaque assays replicated ≥ 2 times with similar results.
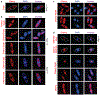
Figure 2:. CRISPR-Cas and restriction proteins are excluded from ΦKZ’s nucleus-like structure.
a, Fluorescence microscopy of P. aeruginosa immunostained for Cas9, DAPI stain shows the phage DNA within the nucleus-like structure. Live fluorescence microscopy of P. aeruginosa strains engineered to express b, II-A Cas9 or I-C or I-F Cas8 or Cas3 proteins fused to Cherry, c, a Cherry-HsdR fusion, d, Immunostained for Myc-ORF152 (top panels), or live imaging of ORF152 and TopA proteins fused to Cherry, or Cherry alone. All experiments were replicated ≥ 2 times with similar results.
Figure 3:. ΦKZ genomic DNA can be cleaved by immune enzymes.
a, ΦKZ and JBD30 genomic DNA digested with the indicated restriction enzymes in vitro. The first lane contains a 1 kb DNA ladder. The number of cut sites for each enzyme is shown at the bottom of the gels. Products were visualized on a 0.7% agarose gel, visualized with SYBR Safe nucleic acid stain. b, ΦKZ phage genomic DNA digested in vitro using KasI, and incubated with and without Cas9 loaded with crRNA:tracrRNA targeting the fragment liberated by KasI. Products were visualized on a 0.7% agarose gel, visualized with SYBR Safe nucleic acid stain. c, Strain PAO1 expressing EcoRI-Cherry, EcoRI-Cherry-ORF152, or EcoRI E111G-Cherry-ORF152 fusion protein. Plaque assays were conducted as in Fig. 1a and quantified (n=3). Mean values are plotted as bars with error bars representing standard deviation. A t-test comparing ΦKZ EOP on PAO1 pEcoRI-Cherry to PAO1 pEcoRI-Cherry-ORF152 yielded a p-value of 2.88×10−4. d, Growth curves monitoring the OD600 of PAO1 cells infected with the indicated ΦKZ multiplicity of infection (MOI). e, Live fluorescence imaging of infected P. aeruginosa strains engineered to express EcoRI-Cherry, EcoRI E111G-Cherry, EcoRI-Cherry-ORF152 or EcoRI E111G-Cherry-ORF152. DAPI stain shows the phage DNA. Cherry shows EcoRI fusion protein. In vitro digestion experiments a and b replicated twice with similar results. Plaque assays (c), growth curves (d), and microscopy (e) were replicated ≥ 3 times.
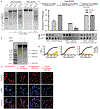
Figure 4:. Phage ΦKZ DNA is sensitive to RNA-targeting Cas13.
a, PAO1 expressing LseCas13a and a crRNA targeting phage ΦKZ. Plaque assays were conducted as in Fig. 1a. b, Growth curves measuring the OD600 of PAO1 infected with the indicated ΦKZ MOI. c, Cas13 escaper ΦKZ phage mutant deletions (red dashed lines) with target sites indicated (blue line). d, Live fluorescence imaging of P. aeruginosa strains engineered to express LseCas13a and crRNAs targeting ΦKZ. Cyan stain shows the phage DNA. e, Model summarizing the ΦKZ nucleus-like structure excluding (flat arrow) Cas9, Cas12, Cascade-Cas3 (Type I-C, Type I-F) and Type I restriction endonucleases (REase) and Type II REase, while the mRNA (red) is exported and can be targeted by Cas13. The nucleus-like structure is resistant to the indicated nucleases, but EcoRI fusion (to internal protein ORF152) enables targeting. Cas13 plaque assays a were replicated >3 times with similar results. Growth curve experiments b were replicated twice with similar results. Escaper mutants c were isolated once and verified by PCR, sequencing, and plaque assays. Microscopy d was replicated twice with similar results.
Comment in
- CRISPR Shields: Fending Off Diverse Cas Nucleases with Nucleus-like Structures.
Barrangou R, Sontheimer EJ. Barrangou R, et al. Mol Cell. 2020 Mar 5;77(5):934-936. doi: 10.1016/j.molcel.2020.02.015. Mol Cell. 2020. PMID: 32142691
Similar articles
- Intracellular Organization by Jumbo Bacteriophages.
Guan J, Bondy-Denomy J. Guan J, et al. J Bacteriol. 2020 Dec 18;203(2):e00362-20. doi: 10.1128/JB.00362-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 32868402 Free PMC article. Review. - A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity.
Malone LM, Warring SL, Jackson SA, Warnecke C, Gardner PP, Gumy LF, Fineran PC. Malone LM, et al. Nat Microbiol. 2020 Jan;5(1):48-55. doi: 10.1038/s41564-019-0612-5. Epub 2019 Dec 9. Nat Microbiol. 2020. PMID: 31819217 - Characterization of a lipid-based jumbo phage compartment as a hub for early phage infection.
Mozumdar D, Fossati A, Stevenson E, Guan J, Nieweglowska E, Rao S, Agard D, Swaney DL, Bondy-Denomy J. Mozumdar D, et al. Cell Host Microbe. 2024 Jul 10;32(7):1050-1058.e7. doi: 10.1016/j.chom.2024.05.016. Epub 2024 Jun 12. Cell Host Microbe. 2024. PMID: 38870941 - Myoviridae bacteriophages of Pseudomonas aeruginosa: a long and complex evolutionary pathway.
Krylov V, Pleteneva E, Bourkaltseva M, Shaburova O, Volckaert G, Sykilinda N, Kurochkina L, Mesyanzhinov V. Krylov V, et al. Res Microbiol. 2003 May;154(4):269-75. doi: 10.1016/S0923-2508(03)00070-6. Res Microbiol. 2003. PMID: 12798231 - Phage phiKZ-The First of Giants.
Krylov V, Bourkaltseva M, Pleteneva E, Shaburova O, Krylov S, Karaulov A, Zhavoronok S, Svitich O, Zverev V. Krylov V, et al. Viruses. 2021 Jan 20;13(2):149. doi: 10.3390/v13020149. Viruses. 2021. PMID: 33498475 Free PMC article. Review.
Cited by
- Intracellular Organization by Jumbo Bacteriophages.
Guan J, Bondy-Denomy J. Guan J, et al. J Bacteriol. 2020 Dec 18;203(2):e00362-20. doi: 10.1128/JB.00362-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 32868402 Free PMC article. Review. - The Dynamics of Synthesis and Localization of Jumbo Phage RNA Polymerases inside Infected Cells.
Antonova D, Belousova VV, Zhivkoplias E, Sobinina M, Artamonova T, Vishnyakov IE, Kurdyumova I, Arseniev A, Morozova N, Severinov K, Khodorkovskii M, Yakunina MV. Antonova D, et al. Viruses. 2023 Oct 16;15(10):2096. doi: 10.3390/v15102096. Viruses. 2023. PMID: 37896872 Free PMC article. - Clades of huge phages from across Earth's ecosystems.
Al-Shayeb B, Sachdeva R, Chen LX, Ward F, Munk P, Devoto A, Castelle CJ, Olm MR, Bouma-Gregson K, Amano Y, He C, Méheust R, Brooks B, Thomas A, Lavy A, Matheus-Carnevali P, Sun C, Goltsman DSA, Borton MA, Sharrar A, Jaffe AL, Nelson TC, Kantor R, Keren R, Lane KR, Farag IF, Lei S, Finstad K, Amundson R, Anantharaman K, Zhou J, Probst AJ, Power ME, Tringe SG, Li WJ, Wrighton K, Harrison S, Morowitz M, Relman DA, Doudna JA, Lehours AC, Warren L, Cate JHD, Santini JM, Banfield JF. Al-Shayeb B, et al. Nature. 2020 Feb;578(7795):425-431. doi: 10.1038/s41586-020-2007-4. Epub 2020 Feb 12. Nature. 2020. PMID: 32051592 Free PMC article. - Identification of the bacteriophage nucleus protein interaction network.
Enustun E, Deep A, Gu Y, Nguyen KT, Chaikeeratisak V, Armbruster E, Ghassemian M, Villa E, Pogliano J, Corbett KD. Enustun E, et al. bioRxiv [Preprint]. 2023 May 18:2023.05.18.541317. doi: 10.1101/2023.05.18.541317. bioRxiv. 2023. PMID: 37292858 Free PMC article. Updated. Preprint. - Accumulation of defense systems in phage-resistant strains of Pseudomonas aeruginosa.
Costa AR, van den Berg DF, Esser JQ, Muralidharan A, van den Bossche H, Bonilla BE, van der Steen BA, Haagsma AC, Fluit AC, Nobrega FL, Haas PJ, Brouns SJJ. Costa AR, et al. Sci Adv. 2024 Feb 23;10(8):eadj0341. doi: 10.1126/sciadv.adj0341. Epub 2024 Feb 23. Sci Adv. 2024. PMID: 38394193 Free PMC article.
References
- Pawluk A et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nature Microbiology 1, 1–6 (2016). - PubMed
METHODS REFERENCES
- Choi K-H & Schweizer HP mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1, 153–161 (2006). - PubMed
- Mesyanzhinov VV et al. The genome of bacteriophage phi KZ of Pseudomonas aeruginosa. Journal of Molecular Biology 317, 1–19 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007810/GM/NIGMS NIH HHS/United States
- T32 AI060537/AI/NIAID NIH HHS/United States
- R01 GM127489/GM/NIGMS NIH HHS/United States
- DP5 OD021344/OD/NIH HHS/United States
- R01 GM104556/GM/NIGMS NIH HHS/United States
- R01 GM129245/GM/NIGMS NIH HHS/United States
- R35 GM118099/GM/NIGMS NIH HHS/United States
- GM104556/Natiional Institutes of Health/International
LinkOut - more resources
Full Text Sources
Research Materials