SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection - PubMed (original) (raw)
. 2019 Dec 18;10(1):5770.
doi: 10.1038/s41467-019-13659-4.
Daniela Niemeyer 3 4, Doreen Muth 3 4, Victor M Corman 3 4, Silvia Martinelli 5, Alwine Gassen 6, Kathrin Hafner 5, Jan Papies 3 4, Kirstin Mösbauer 3 4, Andreas Zellner 5, Anthony S Zannas 5 7 8 9, Alexander Herrmann 10, Florian Holsboer 5, Ruth Brack-Werner 10, Michael Boshart 6, Bertram Müller-Myhsok 5 11 12, Christian Drosten 3 4, Marcel A Müller 3 4 13, Theo Rein 14 15
Affiliations
- PMID: 31852899
- PMCID: PMC6920372
- DOI: 10.1038/s41467-019-13659-4
SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection
Nils C Gassen et al. Nat Commun. 2019.
Abstract
Autophagy is an essential cellular process affecting virus infections and other diseases and Beclin1 (BECN1) is one of its key regulators. Here, we identified S-phase kinase-associated protein 2 (SKP2) as E3 ligase that executes lysine-48-linked poly-ubiquitination of BECN1, thus promoting its proteasomal degradation. SKP2 activity is regulated by phosphorylation in a hetero-complex involving FKBP51, PHLPP, AKT1, and BECN1. Genetic or pharmacological inhibition of SKP2 decreases BECN1 ubiquitination, decreases BECN1 degradation and enhances autophagic flux. Middle East respiratory syndrome coronavirus (MERS-CoV) multiplication results in reduced BECN1 levels and blocks the fusion of autophagosomes and lysosomes. Inhibitors of SKP2 not only enhance autophagy but also reduce the replication of MERS-CoV up to 28,000-fold. The SKP2-BECN1 link constitutes a promising target for host-directed antiviral drugs and possibly other autophagy-sensitive conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures
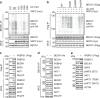
Fig. 1. FKBP51 increases BECN1 stability.
a FKBP51 increases BECN1 protein levels. HEK293 cells were transfected with either Flag-tagged FKBP51 or FKBP52 expressing plasmid or control vector. BECN1 mRNA and protein were determined after 72 h. b BECN1 is subject to proteasomal degradation. HEK293 cells were transfected with Ubiquitin-HA expressing plasmid and treated with the proteasome inhibitor MG132 (10 µM, 2 h) as indicated. BECN1 was immunoprecipitated from whole cell extracts and probed for ubiquitination by western blotting (quantification Supplementary Fig. 1a). c–f FKBP51 delays degradation of BECN1. HEK293 cells were transfected with vector control (Co) or FKBP51 expressing plasmid for 72 h and treated with the translation inhibitor cycloheximide (CHX, 30 µg mL−1) as indicated in c, d. In e, f, HEK293 cells were transfected with Halo-tagged BECN1 expressing plasmid together with FKBP51-Flag expressing plasmid or vector, cultivated with halogenated dye (100 nM, R110Direct, pulse) overnight, switched to medium without dye (chase) and treated with MG132 or vehicle for the indicated times. In all panels, error bars denote the standard error of the mean, derived from n = 3 biologically independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (one-way ANOVA in a, two-way ANOVA in d, f details in Supplementary Table 1). Source data and blot collections are provided as a Source Data file.
Fig. 2. FKBP51 associates with SKP2 that regulates BECN1 stability.
a HEK293 cells were transfected with plasmids expressing the indicated E3 ligases and USPs; after 72 h, cells were either harvested for immunoprecipitation of FKBP51. b BECN1 associates with SKP2, but not with USP18 or USP36. HEK293 cells were transfected with vectors expressing tagged SKP2, USP18 and USP36; after 72 h, cells were harvested for immune-precipitation with either control IgG or anti BECN1 antibody. c, d SKP2 affects BECN1 stability. HEK293 cells were transfected with plasmids expressing SKP2 or TRAF6 and exposed to MG132 (10 µM, 2 h) where indicated and to cycloheximide (CHX, 30 µg mL−1) after 72 h for the durations indicated to monitor the decay of BECN1. e, f The conditions of c,d were also used in the pulse-chase assay, performed as in Fig. 1e, f, to determine BECN1 stability. g, h Cellular SKP2 levels determine BECN1 levels. SKP2 was either down-regulated by using siRNA (g) or up-regulated by transient transfection (h) and the levels of the indicated proteins were measured by western blotting (P27 served as control). Quantifications for g and h are provided in Supplementary Fig. 1c, d. i Knock-down of SKP2 by siRNA decreases BECN1 ubiquitination (assay as in Fig. 1b). j–m Stability assays were performed like in c–f to determine the effect of SKP2-targeting siRNA on BECN1 stability. WCE = whole cell extract. In all panels, error bars denote the standard error of the mean, derived from n = 3 biologically independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (two-way ANOVAs, details in Supplementary Table 1). Source data and blot collections are provided as a Source Data file.
Fig. 3. SKP2 executes K48-linked poly-ubiquitination at K402 of BECN1.
a, b SKP2 attaches K48-linked poly-ubiquitin at K402 of BECN1. HEK293 cells were transfected with plasmids expressing either WT HA-tagged ubiquitin or mutant forms featuring only one lysine together with an SKP2 expressing plasmid or vector control (a). In b, cells were transfected with plasmids expressing BECN1, either WT or the indicated mutants, myc-SKP2 and HA-ubiquitin. Ubiquitination of BECN1 was determined after immunoprecipitation. c–e Association of FKBP51 and BECN1 with regulatory kinases. HEK293 cells were transfected with FKBP51-Flag expressing plasmid (c, e) and protein extracts were used for western blot analysis after 72 h of cultivation (e) or immunoprecipitation of FKBP51 (c). In d, endogenous BECN1 was precipitated from cell extracts. Blot collections are provided as a Source Data file.
Fig. 4. SKP2 is controlled by AKT-PHLPP mediated phosphorylation involving FKBP51.
a Model of FKBP51`s kinase associations that regulate SKP2 phosphorylation and activity. The recruitment of PHLPP by FKBP51 leads to lower phosphorylation (S473) and activity of AKT1, which causes decreased phosphorylation and activity of SKP2, thereby stabilizing BECN1. b, c BECN1 stability was assessed by the cycloheximide assay (as in Fig. 1c, d) comparing wt SKP2 with the phosphor-null mutant S72A and the phosphomimetic mutant S72D/S75D. d–f PHLPP inhibition destabilizes BECN1. HEK293 cells were treated with PHLPPi (NSC117079, 50 µM) for 1 h and cell extracts were probed for the indicated proteins by western blot analysis (d). The pulse-chase assay as in Fig. 1e, f was used in e, f. g–j The cycloheximide protein stability assay was performed (as in Fig. 1c, d) to evaluate the effect of PHLPPi on K402-BECN1 in comparison to wt BECN1 (g, h) and to test the effects of AKT1 inhibitors (AktiX and MK2206) on wt BECN1 (i, j). k, l The effect of the AKT1 inhibitors on BECN1 was also tested in the pulse chase assay. m Reduction of SKP2 by siRNA enhances the half-life of long-lived proteins. HEK293 cells were metabolically labelled with [14C]-valine overnight, chased with fresh media with excess valine overnight and then incubated for 4 h with complete media. The autophagy inhibitor 3-MA (10 mM) was used as control. In all panels, error bars denote the standard error of the mean, derived from n = 3 biologically independent experiments. WCE = whole cell extract. *p < 0.05, **,$$_p_ < 0.01, ***_p_ < 0.001 (two-way ANOVAs, details in Supplementary Table 1. In **m**, **labels the effect of 3-MA in the si-Skp2 RNA condition, labels the effect of the si-Skp2 RNA in the vehicle control. Source data and blot collections are provided as a Source Data file.
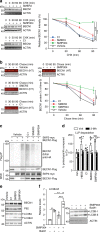
Fig. 5. SKP2 inhibitors enhance BECN1 protein stability and autophagy.
a, b Evaluation of small compounds inhibiting SKP2. HEK293 cells were exposed to known SKP2 inhibitors for 24 h, followed by additional exposure to cycloheximide (CHX, 30 µg mL−1) for the times as indicated or vehicle (a). b HEK293 cells were transfected with Halo-tagged BECN1 expressing plasmid and exposed to known SKP2 inhibitors and halogenated dye (100 nM, R110Direct) overnight. Cells were transferred to medium without dye and harvested after the indicated times. Labelled BECN1 was determined and quantified. c HEK293 cells were transfected with control vector (−) or plasmids expressing BECN1-Flag, HA-ubiquitin and myc-SKP2. Cells were exposed to known SKP2 inhibitors (C1, 3 µM; SMER3, 5 µM; SMIP004, 10 µM) for 24 h, BECN1 was precipitated from cell extracts and its ubiquitination and levels were determined by western blotting. d SKP2 inhibitors impact long-lived proteins. HEK293 cells were metabolically labelled with [14C]-valine overnight, exposed to the indicated drugs or starvation (HBSS = Hank’s balanced salt solution) and the degradation of proteins was determined 4 h after withdrawal of [14C]-valine completed media. The autophagy inhibitor 3-MA (10 mM) was used as control. e, f Assessment of autophagy markers upon exposure to SKP2 inhibitors. VeroB4 cells were exposed to the indicated drugs or vehicle and the levels of P62 and LC3B-II/I were determined by western blotting (e). The most efficient drug, SMIP004, was analyzed in the flux assay with bafilomycin A1 (BafA1; f). In all panels, error bars denote the standard error of the mean, derived from n = 3 biologically independent experiments. WCE = whole cell extract. *,$p < 0.05, **,$$p < 0.01, ***,$$$p < 0.001 (two-way ANOVAs, details in Supplementary Table 1). In d, *labels refer to the effects of 3-MA, $labels to the drug effects in comparison to vehicle. Source data and blot collections are provided as a Source Data file.
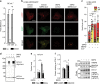
Fig. 6. MERS-CoV blocks autophagic flux.
a MERS-CoV decreases BECN1 and increases pSKP2. VeroB4 cells were infected with MERS-CoV (MOI = 0.001) or mock-infected and harvested 48 h later. Representative western blots are shown, quantification in Supplementary Fig. 3a, b. b MERS-CoV increases K48-linked poly-ubiquitination of BECN1. BECN1 was immune-precipitated from cell extracts 48 h p.i. and its ubiquitination determined by a ubiquitin K48 linkage specific antibody. c, d MERS-CoV blocks fusion of AP with lysosomes. VeroB4 cells were transfected with tandem fluorescent-tagged LC3B (mRFP and EGFP) and infected with MERS-CoV (MOI = 0.001). Twenty-four hours later, cells were fixed and analyzed for fluorescence. Vesicles with both green and red fluorescence (autophagosomes, AP) and with red fluorescence only (autolysosomes, AL) were counted. d, representative images, scale bar 25 µm. e, f MERS-CoV decreases ATG14 oligomerization. VeroB4 cells were infected with MERS-CoV (MOI = 0.001), cross-linked with disuccinimidyl suberate (DSS, 75 µM) 48 h p.i. for 30 min and harvested. ATG14 homo-oligomerization was examined after western blotting (ProteinSimple). g MERS-CoV decreases autophagic flux. VeroB4 cells were infected with MERS-CoV (MOI = 0.001) and incubated with bafilomycin A1 (BafA1, 0.1 µM) for 2 h before samples were taken at 48 h p.i.. The ratios of LC3B-II/I were determined by western blotting. h MERS-CoV affects SNARE protein interactions. VeroB4 cells were infected with MERS-CoV (MOI = 0.001), cross-linked with disuccinimidyl suberate (DSS, 75 µM) 48 h p.i. for 30 min and harvested. The SNARE complex protein STX17 was immunoprecipitated and the eluate was probed for interacting VAMP8 and SNAP29 by western blotting (the quantification represents the bands detected at the combined molecular weights of the cross-linked proteins, i.e. STX17 + VAMP8 and STX17 + SNAP29). In all panels, error bars denote the standard error of the mean. derived from n = 3 biologically independent experiments for b, f, g, h, and n = 13 (control) or n = 12 (CoV) different cells for c. WCE = whole cell extract of vehicle exposed cells, i.e. no cross-linking. *p < 0.05, **p < 0.01, ***p < 0.001 (_t_-test in b, c, f, h, two-way ANOVA in g, details in Supplementary Tables 1, 2). Source data and blot collections are provided as a Source Data file.
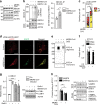
Fig. 7. Mutual influence of MERS-CoV and autophagy.
a, b Deletion of ATG5 in VeroB4 cells facilitates MERS-CoV replication. VeroB4 wt or Atg5 knockout cells were infected with MERS-CoV (MOI = 0.001). Plaque forming units (PFU, a) and genome equivalents (GE, b) per ml were determined by plaque assay or quantitative real time RT-PCR, at 24 and 48 h p.i.. Fold difference and absolute numbers per ml are displayed. In all panels, error bars denote the standard error of the mean, derived from n = 3 biologically independent experiments. **p < 0.01, ***p < 0.001 (_t_-tests, details in Supplementary Table 2). c–f MERS-CoV NSP4, NSP6 (c, d) p3, p4a, p4b, and p5 (e, f, all proteins were Flag-tagged at the N-terminus) were transiently expressed in VeroB4 cells and the indicated proteins were determined by western blotting after 72 h. BafA1 (0.1 µM) was added 2 h before harvesting to assess the autophagic flux (d, f). Western blots are representative of three independent experiments. Source data and blot collections are provided as a Source Data file.
Fig. 8. SKP2 inhibition reduces replication of MERS-CoV and its effects on autophagy.
a The SKP2 inhibitor (SKP2i) efficiently inhibits MERS-CoV replication. VeroB4 cells were infected with MERS-CoV (MOI = 0.001) and treated with SKP2i (10 µM) or DMSO (Co = control). MERS-CoV GE were determined by real-time RT-PCR at 24 and 48 h p.i., data are presented as fold difference (black bars, SKP2i condition) in comparison to control (gray bars, Co = DMSO condition). Raw data presenting GE ml^-1 is shown in Fig. S5A. b, c The SKP2-inhibitor (SKP2i) restores autophagic flux in MERS-CoV-infected cells. VeroB4 cells were transfected with mRFP-EGFP-tagged LC3B, infected with MERS-CoV (MOI = 0.001) and treated with SKP2i (10 µM) or vehicle for 24 h. b Representative images (scale bar 25 µm). c The numbers of vesicles with both green and red fluorescence (autophagosomes, AP) and with red fluorescence only (autolysosomes, AL) were counted 24 h p.i.. d–f SKP2i enhances protein interactions indicative of autolysosome formation in MERS-CoV-infected cells. VeroB4 cells were infected with MERS-CoV (MOI = 0.001), treated with SKP2i for 48 h, cross-linked with disuccinimidyl suberate (DSS, 75 µM) 48 h p.i. for 30 min and harvested. ATG14 homo-oligomerization was assessed after western blotting (d) and quantified (e). f The SNARE complex protein STX17 was immunoprecipitated and the eluate was probed for interacting VAMP8 and SNAP29 by western blotting and quantified as in Fig. 6h. In all panels, error bars denote the standard error of the mean, derived from n = 3 (a, e) or n = 4 (f) biologically independent experiments, and n = 13 (c, vehicle, non-infected) or n = 12 (all other conditions in c) different cells. *p < 0.05, **p < 0.01, ***,###p < 0.001 (c, *,***refer to the statistical difference between the numbers autolysosomes, ###to the difference between the total numbers of fluorescing vesicles). Two-way ANOVA was performed in c, _t_-tests in a, e, f, details in Supplementary Tables 1, 2. Source data and blot collections are provided as a Source Data file.
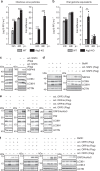
Fig. 9. Some potential SKP2 inhibitors affect autophagy and viral replication.
a VeroB4 cells were treated with various FDA-approved drugs (for abbreviations and concentrations see Supplementary Table 3) and cotreatment with Bafilomycin A1 (0.1 μM) was performed to evaluate the autophagic flux. The graph provides the means + SEM of three independent experiments, representative western blots are shown in Supplementary Fig. 8b. b VeroB4 cells were infected with MERS-CoV (MOI = 0.001), treated with the indicated drugs, and MERS-CoV genome copies were determined by RT-PCR 24 h and 48 h p.i.; data are presented as (log10) fold difference in relation to the DMSO control. Unprocessed data and PFU results are presented in Supplementary Fig. 9a–c. c Concentration-dependent inhibition of MERS-CoV replication by the SKP2-inhibitor (SKP2i), valinomycin (VAL) und niclosamide (NIC). VeroB4 cells were infected with MERS-CoV (MOI = 0.001) and treated with drug. MERS-CoV genome copies were determined by real-time RT-PCR at 48 h p.i., data present the percentage of remaining MERS-CoV genomes in comparison to vehicle treatment. In all panels, error bars denote the standard error of the mean, derived from n = 3 or n = 6 (c, no drug condition) biologically independent experiments. **p < 0.01, ***p < 0.001 (two-way ANOVA in a, one-way ANOVA in b, details in Supplementary Table 1). d Summary scheme: SKP2 leads to K48-linked poly-ubiquitination and thus degradation of BECN1. The effect of SKP2 can be diminished in two ways, either by chemical inhibition or by FKBP51, which scaffolds protein interactions ultimately leading to SKP2 inactivation through inhibiting its phosphorylation. Both scenarios enhance autophagy, which involves ATG14 oligomerization (probably 7–8mers) as recently described essential step in functional autophagy. MERS-CoV reduces autophagy through distinct viral proteins leading to blockade of autophagosome-lysosome fusion and ATG14 oligomerization. Compounds inhibiting SKP2 reinstate autophagy and efficiently reduce viral production. Source data are provided as a Source Data file.
Similar articles
- MicroRNA-205-5p targets E2F1 to promote autophagy and inhibit pulmonary fibrosis in silicosis through impairing SKP2-mediated Beclin1 ubiquitination.
Qian Q, Ma Q, Wang B, Qian Q, Zhao C, Feng F, Dong X. Qian Q, et al. J Cell Mol Med. 2021 Oct;25(19):9214-9227. doi: 10.1111/jcmm.16825. Epub 2021 Aug 24. J Cell Mol Med. 2021. PMID: 34428336 Free PMC article. - UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b.
Zhou Y, Zheng R, Liu D, Liu S, Disoma C, Li S, Liao Y, Chen Z, Du A, Dong Z, Zhang Y, Liu P, Razzaq A, Chen D, Chen X, Zhong X, Liu S, Tao S, Liu Y, Xu L, Deng X, Li J, Jiang T, Zhao J, Li S, Xia Z. Zhou Y, et al. J Virol. 2022 Sep 14;96(17):e0074122. doi: 10.1128/jvi.00741-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980206 Free PMC article. - Ca2+ Ions Promote Fusion of Middle East Respiratory Syndrome Coronavirus with Host Cells and Increase Infectivity.
Straus MR, Tang T, Lai AL, Flegel A, Bidon M, Freed JH, Daniel S, Whittaker GR. Straus MR, et al. J Virol. 2020 Jun 16;94(13):e00426-20. doi: 10.1128/JVI.00426-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295925 Free PMC article. - The BECN1-USP19 axis plays a role in the crosstalk between autophagy and antiviral immune responses.
Cui J, Jin S, Wang RF. Cui J, et al. Autophagy. 2016 Jul 2;12(7):1210-1. doi: 10.1080/15548627.2016.1173801. Epub 2016 Apr 20. Autophagy. 2016. PMID: 27096686 Free PMC article. Review. - Modulation of the immune response by Middle East respiratory syndrome coronavirus.
Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. Shokri S, et al. J Cell Physiol. 2019 Mar;234(3):2143-2151. doi: 10.1002/jcp.27155. Epub 2018 Aug 26. J Cell Physiol. 2019. PMID: 30146782 Free PMC article. Review.
Cited by
- Canonical and Noncanonical Autophagy as Potential Targets for COVID-19.
Bello-Perez M, Sola I, Novoa B, Klionsky DJ, Falco A. Bello-Perez M, et al. Cells. 2020 Jul 5;9(7):1619. doi: 10.3390/cells9071619. Cells. 2020. PMID: 32635598 Free PMC article. Review. - Niclosamide inhalation powder made by thin-film freezing: Multi-dose tolerability and exposure in rats and pharmacokinetics in hamsters.
Jara MO, Warnken ZN, Sahakijpijarn S, Moon C, Maier EY, Christensen DJ, Koleng JJ, Peters JI, Hackman Maier SD, Williams Iii RO. Jara MO, et al. Int J Pharm. 2021 Jun 15;603:120701. doi: 10.1016/j.ijpharm.2021.120701. Epub 2021 May 12. Int J Pharm. 2021. PMID: 33989748 Free PMC article. - Inhibitors of VPS34 and lipid metabolism suppress SARS-CoV-2 replication.
Silvas JA, Jureka AS, Nicolini AM, Chvatal SA, Basler CF. Silvas JA, et al. bioRxiv [Preprint]. 2020 Jul 20:2020.07.18.210211. doi: 10.1101/2020.07.18.210211. bioRxiv. 2020. PMID: 32743584 Free PMC article. Updated. Preprint. - A single-oral bolus of 100,000 IU of cholecalciferol at hospital admission did not improve outcomes in the COVID-19 disease: the COVID-VIT-D-a randomised multicentre international clinical trial.
Cannata-Andía JB, Díaz-Sottolano A, Fernández P, Palomo-Antequera C, Herrero-Puente P, Mouzo R, Carrillo-López N, Panizo S, Ibañez GH, Cusumano CA, Ballarino C, Sánchez-Polo V, Pefaur-Penna J, Maderuelo-Riesco I, Calviño-Varela J, Gómez MD, Gómez-Alonso C, Cunningham J, Naves-Díaz M, Douthat W, Fernández-Martín JL; COVID-VIT-D trial collaborators. Cannata-Andía JB, et al. BMC Med. 2022 Feb 18;20(1):83. doi: 10.1186/s12916-022-02290-8. BMC Med. 2022. PMID: 35177066 Free PMC article. Clinical Trial. - FKBP51 plays an essential role in Akt ubiquitination that requires Hsp90 and PHLPP.
Tufano M, Marrone L, D'Ambrosio C, Di Giacomo V, Urzini S, Xiao Y, Matuozzo M, Scaloni A, Romano MF, Romano S. Tufano M, et al. Cell Death Dis. 2023 Feb 13;14(2):116. doi: 10.1038/s41419-023-05629-y. Cell Death Dis. 2023. PMID: 36781840 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous