Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries - PubMed (original) (raw)
Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries
Matthew R Olm et al. mSystems. 2020.
Abstract
Longstanding questions relate to the existence of naturally distinct bacterial species and genetic approaches to distinguish them. Bacterial genomes in public databases form distinct groups, but these databases are subject to isolation and deposition biases. To avoid these biases, we compared 5,203 bacterial genomes from 1,457 environmental metagenomic samples to test for distinct clouds of diversity and evaluated metrics that could be used to define the species boundary. Bacterial genomes from the human gut, soil, and the ocean all exhibited gaps in whole-genome average nucleotide identities (ANI) near the previously suggested species threshold of 95% ANI. While genome-wide ratios of nonsynonymous and synonymous nucleotide differences (dN/dS) decrease until ANI values approach ∼98%, two methods for estimating homologous recombination approached zero at ∼95% ANI, supporting breakdown of recombination due to sequence divergence as a species-forming force. We evaluated 107 genome-based metrics for their ability to distinguish species when full genomes are not recovered. Full-length 16S rRNA genes were least useful, in part because they were underrecovered from metagenomes. However, many ribosomal proteins displayed both high metagenomic recoverability and species discrimination power. Taken together, our results verify the existence of sequence-discrete microbial species in metagenome-derived genomes and highlight the usefulness of ribosomal genes for gene-level species discrimination.IMPORTANCE There is controversy about whether bacterial diversity is clustered into distinct species groups or exists as a continuum. To address this issue, we analyzed bacterial genome databases and reports from several previous large-scale environment studies and identified clear discrete groups of species-level bacterial diversity in all cases. Genetic analysis further revealed that quasi-sexual reproduction via horizontal gene transfer is likely a key evolutionary force that maintains bacterial species integrity. We next benchmarked over 100 metrics to distinguish these bacterial species from each other and identified several genes encoding ribosomal proteins with high species discrimination power. Overall, the results from this study provide best practices for bacterial species delineation based on genome content and insight into the nature of bacterial species population genetics.
Keywords: bacterial species; bioinformatics; metagenomics; microbial genetics; species.
Copyright © 2020 Olm et al.
Figures
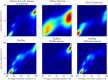
FIG 1
Average nucleotide identity gaps exist near ∼95% ANI in all tested genome sets. Each plot is a histogram of average nucleotide identity and genome alignment percentage values resulting from pairwise comparison within a genome set. Higher-intensity colors represent a higher density of comparisons with that particular ANI and genome alignment percentage. The top row contains data from three sets of metagenome-assembled genomes (MAGs) from different environments. The bottom row displays data from NCBI RefSeq (rarefied to reduce taxonomic bias; see Materials and Methods), RefSeq with only comparisons between genomes annotated as the same species included, and RefSeq with only comparisons between genomes annotated as different species included.
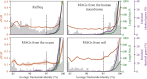
FIG 2
Metrics of recombination and selection follow patterns related to the proposed 95% ANI species threshold. Each plot displays a histogram of ANI values resulting from pairwise comparison within a genome set (light gray bars), the median dN/dS ratio at each ANI level (orange line), and the median estimated recombination rate at each ANI level determined using two criteria, namely, the percentage of enriched identical genes (purple line; see Materials and Methods for details) and length bias (green line), as measured using the program PopCOGent. A dotted line is drawn at 95% ANI to mark the commonly proposed threshold for species delineation, and 95% confidence intervals are shown shaded around orange, green, and purple lines. Color coding corresponds to _y_-axis labels.
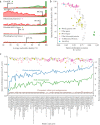
FIG 3
Whole-genome alignment outperforms analysis of marker genes for species discrimination. (a) Histograms of ANI values from comparisons between bacteria from RefSeq annotated as belonging to the same species (green) or different species (red). Each row represents a different method of nucleotide sequence alignment, and vertical black lines indicate the ANI value with the highest F1 score for the corresponding method. (b) Comparison of optimal species discrimination threshold to F1 score for reconstruction of species-level clusters from RefSeq. Whole-genome comparison algorithms, a 16S rRNA alignment, and single-copy gene alignments were tested. (c) Accuracy of marker genes for reconstruction of species clusters based on 95% ANI whole-genome alignments of genomes from metagenomes (dots; left y axis) and recoverability of maker genes from metagenomic data from different environments (lines; right y axis). A horizontal dotted line marks a recoverability level of 1, meaning equal numbers of marker genes and genomes were assembled from the environment.
Similar articles
- Promiscuous and genome-wide recombination underlies the sequence-discrete species of the SAR11 lineage in the deep ocean.
Zhao J, Pachiadaki M, Conrad RE, Hatt JK, Bristow LA, Rodriguez-R LM, Rossello-Mora R, Stewart FJ, Konstantinidis KT. Zhao J, et al. ISME J. 2025 Apr 18:wraf072. doi: 10.1093/ismejo/wraf072. Online ahead of print. ISME J. 2025. PMID: 40247698 - Exploration of the genetic landscape of bacterial dsDNA viruses reveals an ANI gap amid extensive mosaicism.
Ndovie W, Havránek J, Leconte J, Koszucki J, Chindelevitch L, Adriaenssens EM, Mostowy RJ. Ndovie W, et al. mSystems. 2025 Feb 18;10(2):e0166124. doi: 10.1128/msystems.01661-24. Epub 2025 Jan 29. mSystems. 2025. PMID: 39878503 Free PMC article. - High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries.
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. Jain C, et al. Nat Commun. 2018 Nov 30;9(1):5114. doi: 10.1038/s41467-018-07641-9. Nat Commun. 2018. PMID: 30504855 Free PMC article. - Recovering complete and draft population genomes from metagenome datasets.
Sangwan N, Xia F, Gilbert JA. Sangwan N, et al. Microbiome. 2016 Mar 8;4:8. doi: 10.1186/s40168-016-0154-5. Microbiome. 2016. PMID: 26951112 Free PMC article. Review. - [A review on the bioinformatics pipelines for metagenomic research].
Ye DD, Fan MM, Guan Q, Chen HJ, Ma ZS. Ye DD, et al. Dongwuxue Yanjiu. 2012 Dec;33(6):574-85. doi: 10.3724/SP.J.1141.2012.06574. Dongwuxue Yanjiu. 2012. PMID: 23266976 Review. Chinese.
Cited by
- Cofitness network connectivity determines a fuzzy essential zone in open bacterial pangenome.
Zhang P, Zhang B, Ji YY, Jiao J, Zhang Z, Tian CF. Zhang P, et al. mLife. 2024 Jun 28;3(2):277-290. doi: 10.1002/mlf2.12132. eCollection 2024 Jun. mLife. 2024. PMID: 38948139 Free PMC article. - Evolutionary ecology of microbial populations inhabiting deep sea sediments associated with cold seeps.
Dong X, Peng Y, Wang M, Woods L, Wu W, Wang Y, Xiao X, Li J, Jia K, Greening C, Shao Z, Hubert CRJ. Dong X, et al. Nat Commun. 2023 Feb 28;14(1):1127. doi: 10.1038/s41467-023-36877-3. Nat Commun. 2023. PMID: 36854684 Free PMC article. - Strain-Level Variation and Diverse Host Bacterial Responses in Episymbiotic Saccharibacteria.
Nie J, Utter DR, Kerns KA, Lamont EI, Hendrickson EL, Liu J, Wu T, He X, McLean J, Bor B. Nie J, et al. mSystems. 2022 Apr 26;7(2):e0148821. doi: 10.1128/msystems.01488-21. Epub 2022 Mar 28. mSystems. 2022. PMID: 35343799 Free PMC article. - Microbial communities of Auka hydrothermal sediments shed light on vent biogeography and the evolutionary history of thermophily.
Speth DR, Yu FB, Connon SA, Lim S, Magyar JS, Peña-Salinas ME, Quake SR, Orphan VJ. Speth DR, et al. ISME J. 2022 Jul;16(7):1750-1764. doi: 10.1038/s41396-022-01222-x. Epub 2022 Mar 28. ISME J. 2022. PMID: 35352015 Free PMC article. - Accurate and robust inference of microbial growth dynamics from metagenomic sequencing reveals personalized growth rates.
Joseph TA, Chlenski P, Litman A, Korem T, Pe'er I. Joseph TA, et al. Genome Res. 2022 Mar;32(3):558-568. doi: 10.1101/gr.275533.121. Epub 2022 Jan 5. Genome Res. 2022. PMID: 34987055 Free PMC article.